Figure 8.
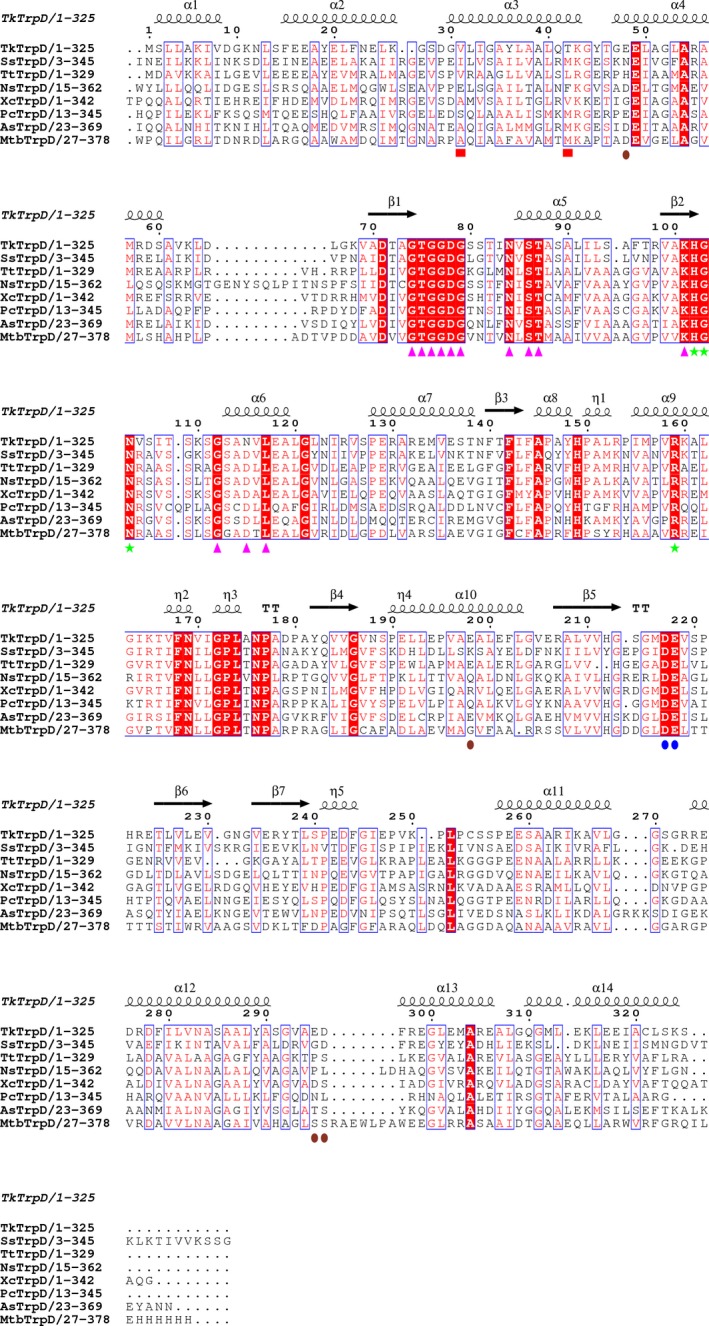
Structure‐based sequence alignment of TkTrpD. The enzymes used are SsTrpD (PDB entry 2GVQ) 7, TtTrpD (PDB entry 1V8G; Shimizu et al., 2004, unpublished), NsTrpD (PDB entry 1VQU; Joint Center for Structural Genomics, 2005, unpublished), XcTrpD (PDB entry 4HKM; Ghosh et al., 2012, unpublished), PcTrpD (PDB entry 1KHD) 8, AsTrpD (PDB entry 4YI7; Evans et al., 2015, unpublished), and MtbTrpD (PDB entry 4X5B) 9. Figure was created using espript 3.0 51. Conserved residues are indicated by white letters on a red background (strictly conserved) or red letters on a white background (global similarity score, 0.7) and framed in blue boxes. Markers indicate residues postulated to be involved in PRPP binding (magenta arrows), anthranilate binding (green stars), metal binding (blue ovals), and dimerization (red boxes). Residues involved in Zn2+ binding at the TrpD–Zn2+ dimer interface are shown with brown ovals.
