Figure 4.
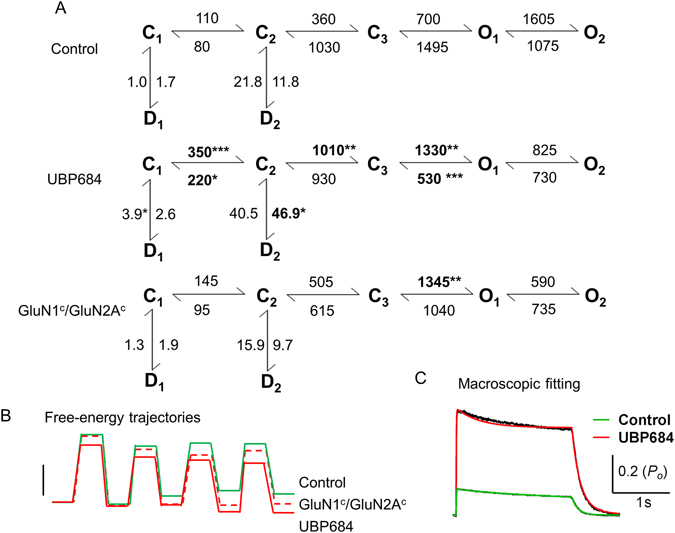
Kinetic mechanism describing the effects of UBP684 on GluN1/GluN2A receptor activation. (A) MIL fit of single-channel data to assess the gating mechanism underlying potentiation of UBP684 is shown. All rates are in sec−1. Bold numbers with asterisks denote the rates which were significantly different from glutamate/glycine control patches. Rates ± SEM are presented in Table 2. Data was analyzed using unpaired t-test. (B) Free-energy trajectories for the kinetic states in the different models are presented. The free energies of the active states for the mutant GluN1c/GluN2Ac is lower than control but is still higher than UBP684. Scale bar represents 1 kBT. (C) Macroscopic current profiles were obtained by whole-cell recordings (black traces). Cells were equilibrated to glycine ± UBP684 before exposing to glycine + glutamate ± UBP684 as detailed in Methods. Current profiles were fitted to the kinetic Markov models for control (green) and UBP684 (red) that included glutamate binding and unbinding steps. Except the desensitization rates and the agonist binding and unbinding rates all other rates were fixed to those depicted in the kinetic models. The models were able to accurately predict the activation and deactivation kinetics. Rates obtained from least squares fitting are presented in Table 3.
