Figure 5.
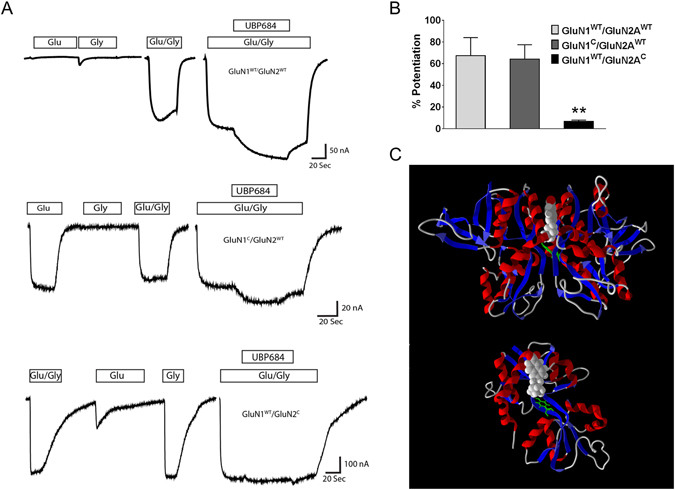
UBP684 potentiation is dependent on GluN2 LBD flexibility. (A) Current traces showing the effect of UBP684 on wildtype, GluN1 LBD-locked (GluN1C/GluN2WT) or GluN2 LBD-locked (GluN1WT/GluN2C) receptors. (B) UBP684 potentiated wildtype and GluN1 LBD-locked receptors but not GluN2 LBD-locked receptors (one-way ANOVA followed by Tukey’s test; P = 0.0012 and 0.0093 for GluN1WT/GluN2Ac compared to GluN1WT/GluN2AWT and GluN1C/GluN2AWT respectively; N = 19–32). (C) Molecular modeling of UBP684 binding to the GluN1/GluN2A receptor LBD dimer. Top: UBP684 (space filled) is shown docked into the GluN1/GluN2A LBD intersubunit interface. Modeling suggests that the carboxylic acid group of UBP684 interacts with positively charged residues on the top of the LBD away from the transmembrane domains and near the N-terminal domains. The alkyl side chain terminates near the LBD hinge region near GluN1 Y535 shown in green. Bottom: The same docking of UBP684 is shown rotated 90° in the horizontal plane with the GluN1 LBD removed and GluN1’s Y535 shown for reference.
