Figure 1.
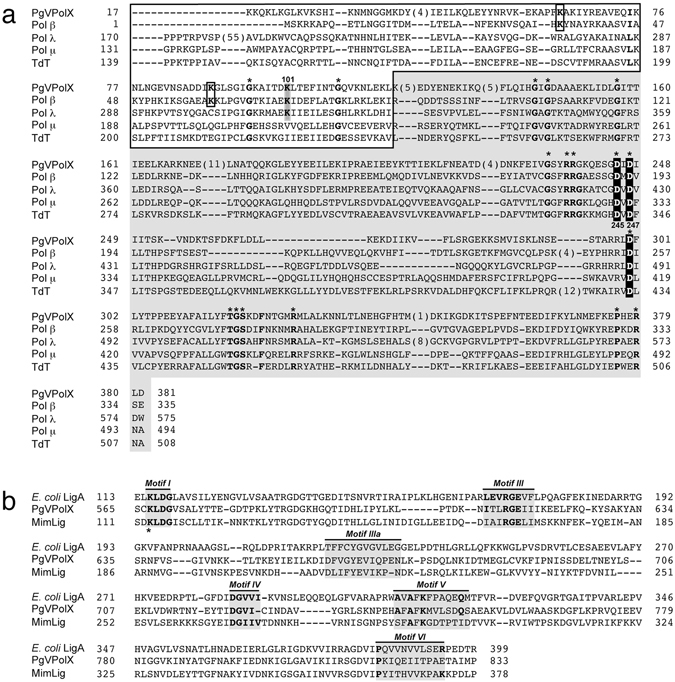
(a) Multiple sequence alignment of the Pol β-like core of PgVPolX with human family X DNA polymerases. Numbers indicate the amino acid position relative to the N-terminus of each DNA polymerase. Names of organisms are abbreviated as follows (numbers in parentheses indicate the corresponding accession number): PgV-16T PolX, DNA polymerase X from strain 16 T of P. globosa virus (NC_021312.1); Polβ, human DNA polymerase β (NP_002681); Pol λ, human DNA polymerase λ (NP_037406); Polμ, human DNA polymerase μ (NP_001271260.1); TdT, terminal deoxynucleotidyl transferase (NP_001017520.1). White letters boxed in black indicate the three conserved Asp responsible for the polymerization reaction. Black bold letters boxed in gray indicate the nucleophile residue responsible for the 5′-dRP lyase activity in Pol β and Pol λ. Black bold letters boxed in white indicate the other two lysine residues described to play a role in 5′-dRP lyase catalysis in Pol β. Asterisks indicate invariant residues among PolX members11. According to Pol β structural data17, 60, the alignment is divided into the 8-kDa subdomain (white area), and the polymerization domain (grey area). (b) Alignment of the C-terminal NAD + -dependent DNA ligase domain of PgVPolX with the Ligase A from E. coli and Mimivirus DNA Ligase (MimLig). The conserved Motifs I-VI present in NAD+-dependent DNA ligases are indicated61. An asterisk indicates the lysine residue that is specifically adenylated during the ligation reaction. The accession numbers of E. coli LigA and MimLig are WP_001413640.1 and Q5UPZ0.1, respectively.
