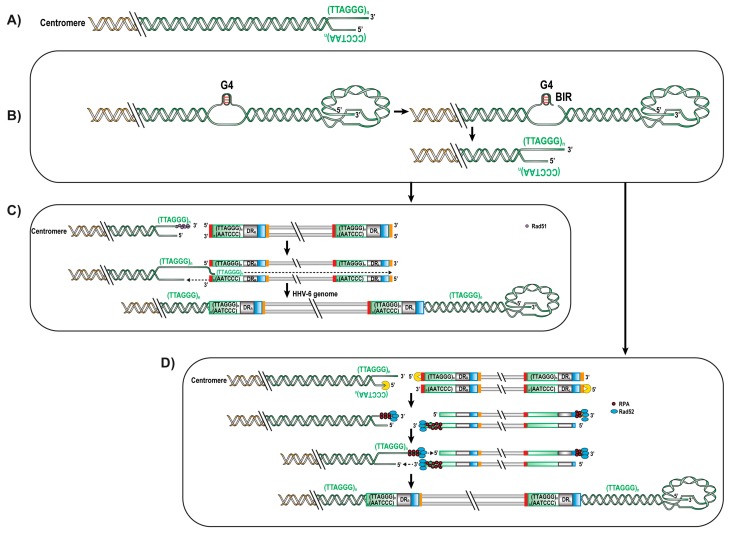
Figure 3.
Possible mechanisms for HHV-6A/B integration. Schematic representation of HHV-6A/B chromosomal integration process. (A) Unfolding of the chromosome t-loop and invasion by the telomeric 3′ overhang into HHV-6A/B’s DR. This mechanism is unlikely to occur since all ciHHV-6A/B reported so far have lost most telomeric repeats. (B) Break induced replication (BIR) repair mechanism caused by G quadruplexes (G4) structure (or other blockage) in the lagging strand. (C) The free 3′ strand is rescued by Rad51 protein that searches for proximal homologous sequences. If a HHV-6A/B genome is close to proximity, Rad51 invades the viral TMR, displacing one strand of the HHV-6A/B genome to allow the synthesis of the complementary strand. Upon cell divisions, the DRL would lose pac1 due to end replication problem and the impTMR would serve as telomeric template to elongate telomeres at the end of the genome. (D) Single stranded annealing (SSA) repair mechanism. Upon virus entry in the cell, DNA damage response is triggered, at the same time a break caused by a stalled replication fork at the human telomeres activate SSA. SSA activation leads to resection of both the viral and human DNA in a 5′ to 3′ direction to create complementary sequences. Meanwhile, the 3′ strands of both genomes are protected by the replication protein A (RPA). Rasd52 binds the RPA and searched for pairing in which pac2 will be lost. Annealed sequences then lead to the copying of the viral genome. Genomes are not drawn to scale.