Figure 6.
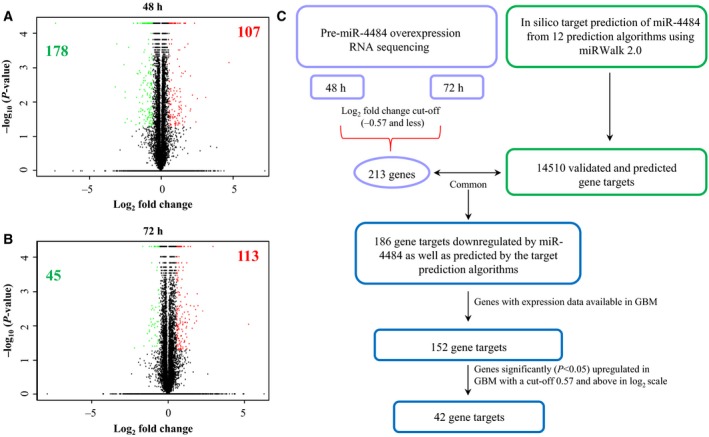
Transcriptome profile upon miR‐4484 overexpression. (A,B) Volcano plot representation of the RNA sequencing data, indicating the genes significantly and differentially expressed between pre‐miR‐neg and pre‐miR‐4484 condition in LN229 cell line at two 48‐ and 72‐h time points, respectively. Each red dot indicates a gene significantly upregulated in pre‐miR‐4484 overexpression condition, and each green dot indicates a gene significantly downregulated in pre‐miR‐4484 overexpression condition. The figures (bolded) inside each plot denote the number of genes upregulated and downregulated significantly. Green indicates the downregulated genes, and red indicates the upregulated genes. (C) Schematic representation of miRNA target prediction. All the genes downregulated (−0.57 and less in the log2 scale) upon miR‐4484 overexpression from both 48‐ and 72‐h time point were sought for the analysis. miR‐4484 targets were derived from 12 target prediction algorithms (miRWalk, MicroT4, miRanda, miRBridge, miRDB, miRMap, miRNAMap, PICTAR2, PITA, RNA22, RNAhybrid and Targetscan) assembled in miRWalk 2.0 online tool. The experimental targets were compared with the in silico targets to find out the common ones, which were finally compared for their expression in GBM.
