Abstract
Bipolar disorder (BD), particularly BD II, is frequently misdiagnosed as unipolar depression (UD), leading to inappropriate treatment and poor clinical outcomes. Although depressive symptoms may be expressed similarly in UD and BD, the similarities and differences in the architecture of brain functional networks between the two disorders are still unknown. In this study, we hypothesized that UD and BD II patients would show convergent and divergent patterns of disrupted topological organization of the functional connectome, especially in the default mode network (DMN) and the limbic network. Brain resting-state functional magnetic resonance imaging (fMRI) data were acquired from 32 UD-unmedicated patients, 31 unmedicated BD II patients (current episode depressed) and 43 healthy subjects. Using graph theory, we systematically studied the topological organization of their whole-brain functional networks at the following three levels: whole brain, modularity and node. First, both the UD and BD II patients showed increased characteristic path length and decreased global efficiency compared with the controls. Second, both the UD and BD II patients showed disrupted intramodular connectivity within the DMN and limbic system network. Third, decreased nodal characteristics (nodal strength and nodal efficiency) were found predominantly in brain regions in the DMN, limbic network and cerebellum of both the UD and BD II patients, whereas differences between the UD and BD II patients in the nodal characteristics were also observed in the precuneus and temporal pole. Convergent deficits in the topological organization of the whole brain, DMN and limbic networks may reflect overlapping pathophysiological processes in unipolar and bipolar depression. Our discovery of divergent regional connectivity that supports emotion processing could help to identify biomarkers that will aid in differentiating these disorders.
Introduction
Affective disorders, such as unipolar depression (UD, or major depressive disorder) and bipolar disorder (BD), are highly prevalent and debilitating conditions associated with high suicide rates and a heavy social burden.1 Although BD consists of recurring episodes of mania/hypomania and depression, the depressive episodes are the most common mood manifestation of the illness.2 Unfortunately, when BD manifests as the depressed state, it is often misdiagnosed as UD, leading to inappropriate treatment, poor clinical outcomes and greater health-care costs.3 It is, therefore, crucial to identify objective and reliable neurobiological markers to help differentiate these disorders, especially in individuals presenting during depressive episodes.
Accumulating evidence suggests widespread brain structural and functional alterations in both UD and BD (mostly BD I), such as the prefrontal cortex, limbic system, ventral striatum, insula and thalamus.4, 5, 6, 7, 8 However, direct comparisons of neuroimaging measures between UD and BD depression are sparse and the results are inconclusive. Structural neuroimaging studies suggest more widespread abnormalities in white matter connectivity and white matter hyperintensities in BD than in UD.3, 6, 9, 10, 11 Task-based functional magnetic resonance imaging (fMRI) studies reported patterns of functional abnormalities in the emotion processing, reward and emotion regulation neural circuits in UD that were distinct from those in BD.3, 4, 6, 12, 13 These abnormalities may help distinguish BD from UD depression. Recently, resting-state fMRI (R-fMRI) has become widely used, as well. By using resting-state functional connectivity (FC) analysis, several studies found FC alterations in the default mode network (DMN) and affective network in different types of affective disorders.5, 8, 14, 15 Moreover, Marchand et al.16 found that the right posterior cingulate cortical FC not only differentiates unipolar from bipolar depression but may also mediate differences in neural processes associated with symptom expression (for example, depression severity and current suicidal ideation) between unipolar and bipolar individuals. However, Anand et al.17 and Wang et al.18 failed to observe a significant difference in the resting-state FC between these two disorders, a finding that might imply an overlap in the neuropathology of depression. However, some of the extant studies included BD- and UD-depressed groups that were not well matched clinically (such as in the duration of illness or the number of episodes, as well as in the levels of depressive symptomatology), some included individuals taking a diverse array of medications and some included both BD I and BD II individuals.6 Notably, there are two established subtypes of BD: type I and type II. The subtype BD II is distinguished from BD I mainly by the absence of full-blown manic episodes. Most neuroimaging studies have involved bipolar I patients, but comparatively few studies have focused on bipolar II disorder.8 However, a growing body of evidence suggests that there may be neurobiological differences between BD I and BD II.19, 20, 21 Therefore, paying attention to the pathophysiology and the neurobiological mechanisms underlying BD II is important.
Graph theory provides a powerful mathematical framework for describing the topological organization of brain networks consisting of nodes (that is, brain regions) and edges (that is, physical or functional connections between brain regions).22 Using graph theoretical methods, researchers revealed that the intrinsic activity of a healthy human brain tends to demonstrate efficient, ‘small-world’ networks that have an optimal balance between local specialization and global integration for information processing.23 Optimal small-world networks have a high level of local clustering (that is, nodes are often connected to their neighbors), combined with short path lengths (that is, it takes few steps from any node to any other node in the network) and a low network cost (that is, the mean physical distance between connected nodes is considerably less than is the case in a random network).23 Several studies have found disrupted small-world networks, modular architecture and abnormal regional characteristics in various brain diseases, such as Alzheimer’s disease,24 attention-deficit/hyperactivity disorder25 and schizophrenia.26 Until now, only a few studies have examined the topology of the whole-brain functional network in UD using R-fMRI and graph theory, and those studies that did were inconsistent in the network topological properties that they reported.27, 28, 29, 30, 31 With respect to BD, only one study showed a disrupted functional topological architecture of the resting-state brain network in BD with hypomanic/manic and depressed episodes (both BD I and II included).32
To our best knowledge, no study has directly compared the topological organization of large-scale whole-brain functional networks between UD and BD II patients. In this study, we acquired R-fMRI data from currently unmedicated UD- and BD II-depressed patients as well as from healthy controls. Then, we analyzed the topological organization of the whole-brain intrinsic functional networks and determined between-group differences in topological parameters. We hypothesized that UD and BD II patients would show convergent and divergent patterns of disrupted topological organization of the functional connectome, especially in the DMN and the limbic network, both of which support emotion and cognitive functioning. Furthermore, we used a receiver operator characteristic (ROC) analysis to investigate whether these topological organizations showed abnormalities which can discriminate between unipolar and bipolar depression.
Materials and methods
Participants
A total of 32 currently depressed patients with bipolar II disorder and 31 currently depressed patients with UD were recruited from the Psychiatry Department of the First Affiliated Hospital of Jinan University, Guangzhou, China. The patients were 18–54 years old. The diagnoses of UD and BD type II were made according to the Structured Clinical Interview for DSM-IV by two experienced psychiatrists. The clinical state for each patient was assessed using the 24-item Hamilton Depression Rating Scale (HAMD) and the Young Mania Rating Scale during the 7-day period prior to the R-fMRI scan. The inclusion criterion for the patients with UD was a total HAMD-24 score >21, whereas for the depressed patients with BD II was a total Young Mania Rating Scale score <7 and total HAMD-24 score >21. The exclusion criteria were patients with other Axis-I psychiatric disorders (except for UD, BD and anxiety disorders), a history of organic brain disorders, neurological disorders, mental retardation, cardiovascular diseases, alcohol/substance abuse or dependence, pregnancy or any physical illness. None of the patients had ever received electroconvulsive therapy prior to participating in the study. In addition, all the patients with UD had no family history of BD. At the time of scanning, all the patients were either medication-naive or had been unmedicated for at least 5 months.
We also recruited 43 healthy subjects as the controls via local advertisements. They were carefully screened using a diagnostic interview, the Structured Clinical Interview for DSM-IV-Nonpatient Edition, to rule out the presence of current or past psychiatric illness. Further exclusion criteria for the healthy controls were any history of psychiatric illness in first-degree relatives and current or past significant medical or neurological illness.
All the subjects were right-handed according to their self-report. In addition, all the included subjects were determined to have no brain abnormalities on conventional MRI by two experienced radiologists. The study was approved by the Ethics Committee of the First Affiliated Hospital of Jinan University, China. Each subject signed a written informed consent form after a full written and verbal explanation of the study.
Data acquisition
All MRI data were obtained on a GE Discovery MR750 3.0 T System with an eight-channel, phased-array head coil. The R-fMRI data were acquired using a single-shot gradient-echo echo planar imaging sequence with the following parameters: repetition time=2000 ms, echo time=25 ms, flip angle=90°, field of view=240 mm × 240 mm, data matrix=64 × 64, thickness/gap=3.0/1.0 mm, 35 axial slices covering the whole brain and 210 volumes acquired in 7 min. During the scanning, each subject was asked to keep their eyes closed, to relax the mind, but not to fall asleep and not to think of anything in particular. In addition, two routine scans using axial T1-weighted fluid attenuation inversion recovery and fast spin-echo T2-weighted MR sequences were also applied to obtain brain images to confirm the absence of any brain structural abnormalities.
Data preprocessing
The R-fMRI data were preprocessed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm/) and DPARSF33 (http://restfmri.net/forum/DPARSF). Preprocessing comprised the removal of the first 10 volumes, slice timing correction for the remaining 200 volumes, realignment to the first volume for head motion correction, spatial normalization to the Montreal Neurological Institute space with 3 × 3 × 3 mm3 voxels, signal linear detrending and temporal band-pass filtering (0.01–0.08 Hz). All the subjects in this study satisfied our criteria for head motion, displacement <1.5 mm in any plane and rotation <1.5° in any direction.
Constructing brain functional networks
Using the GRETNA toolbox34 (http://www.nitrc.org/projects/gretna/), we constructed the brain functional network for each subject according to the automated anatomical labeling (AAL) template.35 First, we randomly parcellated the regions of the AAL template into 1024 ROIs with equal volume size (referred to as the AAL-1024 template) by using the algorithm developed by Zalesky et al.,36 each ROI being defined as a node of the network. For a given ROI in each subject, we obtained its time series by averaging the time courses of all the voxels within the ROI and performed a linear regression to remove the effects of the following nuisance covariates: the Friston-24 parameters of head motion (six head motion parameters, six head motion parameters one time point before and the twelve corresponding squared items)37 and the signals of the whole brain, white matter and cerebrospinal fluid. Second, for each node, we first used the residual of its time series to calculate Pearson’s correlation coefficients with the other 1023 ROIs and then repeated this process for all the ROIs. Thus, we obtained a 1024 by 1024 symmetric correlation matrix for each subject and the corresponding P-value for each matrix element for each subject. Each P-value was the probability of getting a correlation as large as the observed value by random chance when the true correlation is zero. Third, we employed a thresholding procedure on the correlation matrix for each subject using two previous studies.24, 38 Specifically, internodal correlations surviving a threshold of P<0.05 (Bonferroni correction) were retained or, otherwise, were set to 0. Of note, during the calculations, negative correlations were excluded, given their ambiguous interpretation and detrimental effects on test–retest reliability.39, 40, 41, 42 Finally, by taking the remaining Pearson’s coefficients as the edge weights, we obtained a weighted 1024 by 1024 FC matrix for each subject, which was used in the subsequent analyses. We also displayed the mean FC and one subject’s FC pattern from each group as examples to help to clarify the FC matrix (Supplementary Figure S2).
Network analysis
Global parameters
Graph theory was used to analyze the topological properties of the human brain functional networks. We used four parameters, the clustering coefficient (Cw), characteristic path length (Lw), global efficiency (Eglob) and local efficiency (Eloc),43 to characterize the global topological properties of the brain networks. The Cw and Eloc indicate the functional segregation in the brain in that Cw shows the extent of the local interconnectivity or cliquishness in a network and Eloc measures the ability to transfer local information. Lw and Eglob indicate the functional integration in the brain in that Lw measures the harmonic length between pairs and quantifies the ability of information to propagate in parallel and Eglob measures the ability to transfer global information. Their definitions and descriptions are provided in Supplementary Table S1. In addition, we utilized three parameters: the normalized clustering coefficient (γ), normalized weighted characteristic path length (λ) and small worldness (δ) to measure the small-world property. Typically, a small-world network satisfies the following criteria: γ>>1 and λ≈1, or σ=γ/λ>1.44
Modular parameters
We calculated the modularity of the brain network using an optimized algorithm,45 based on the average FC matrix of the healthy controls. Modularity is defined as the extent to which a network can be decomposed into subnetworks that are more connected within modules than between modules. The definition of the modularity index Q is also given in Supplementary Table S1.
On the basis of the detected modules, we calculated the values of the intra- and intermodular FC for each subject. The strength of the intramodular FC was defined as the mean of all the internodal FCs within the selected module, whereas the strength of intermodular FC was defined as the mean of the FCs between any two nodes in the two selected modules. For each module, we also used the intramodular FC to estimate the values of the four global parameters (Cw, Lw, Eloc and Eglob) to characterize the model’s topological properties.
Nodal parameters
We also used two parameters, nodal strength (
) and nodal efficiency (
) to characterize the nodal properties of the brain networks.
quantifies the extent to which a node is relevant to the graph, and Eiw is the ability to transfer information from one node to others. Their definitions and descriptions are also provided in Supplementary Table S1.
Statistical analysis
Group effect analysis
A non-parametric permutation test, which inherently accounts for multiple comparisons,46, 47 was used to detect differences between the UD, BD II and control groups in each of the demographic information, clinical variables, global parameters (Cw, Lw, Eloc, Eglob, γ, λ and δ), intra- and intermodular FCs and nodal parameters (
and
). In the calculations, we took age, gender and age–gender interaction as nuisance covariates and regressed them out. The adjective ‘permutation’ applied to parametric tests (t-tests, analysis of variance) indicates that we permuted the group to determine the P-value of the corresponding t or F statistic rather than using the traditional, Gaussian-based P-values. A detailed description of the permutation test is presented in the Supplementary Materials. The false discovery rate (FDR) method48 was used to correct for multiple comparisons when needed.
Once a significant group effect was observed for a parameter, we estimated its effect size (η2) according to Cohen’s definition,49 according to which the levels of small, medium and large effect size correspond to 0.01, 0.06 and 0.14, respectively. In addition, we performed post hoc tests to determine the between-group differences for each of the network parameters (10 000 permutations).
On the basis of the individual brain functional networks, we also performed a network-based statistic analysis36 to identify any subnetworks, or clusters of connections, in which each edge showed significant differences between the UD patients and the control group or between the BD patients and the control group. This approach can control the family-wise error rate when mass-univariate testing is performed at every connection in the subnetwork.36 The description of the network-based statistic approach is presented in the Supplementary Materials.
Correlations between the network parameters and clinical variables
For the network parameters that showed a significant group effect, we performed a partial correlation analysis to describe their relationships with the clinical variables in each patient group (P<0.05, FDR-corrected). These clinical variables included the HAMD scores, number of episodes, onset age of illness and duration of illness.
Robustness
Robustness is a key issue in network analysis.50, 51 To determine whether the observed group differences in the topological parameters reflect true differences rather than artifacts, we repeated the network analyses using each of the following approaches. First, we used both the positive and negative correlations to construct the brain network. Second, we constructed the binary network. Third, we constructed the network based on a sparsity range. Finally, we considered the effect of the global signal. Specifically, we constructed the network based on the positive correlations and then constructed it based on both the positive and negative correlations without regressing out the global signal.
Network classification
For those network parameters showing a significant group effect, we plotted the ROC curve to determine which of these parameters could clearly distinguish the UD patients from the controls, the BD II patients from the controls and the BD II from the UD patients. The ROC curve is a fundamental plot in signal detection theory52, 53 and is widely used in medical science and neuroimaging studies. A ROC curve, a scatter plot showing the relationship between the false-positive rates and the true-positive rates, describes the relationship between the underlying distribution of the places where signals were absent and the places where signals were present. This analysis was performed using a public MATLAB code (http://www.mathworks.com/matlabcentral/fileexchange/199500-roc-curve; by Giuseppe Cardillo).
Results
Demographic information
Table 1 lists the demographic and clinical characteristics of the three groups. No significant differences were detected between the three groups in age, gender, education levels or the mean framewise displacement parameter. In addition, there was no significant difference in depression severity, illness duration or onset age of illness between the two depressed groups.
Table 1. Demographics and clinical characteristics of the patients with UD, patients with BD and HCs.
| Parameters | UD (n=31) | BD II (n=32) | HC (n=43) | P-value |
|---|---|---|---|---|
| Age (years) | 30.41±9.85 | 28.22±10.13 | 30.19±11.11 | 0.23a |
| Gender | 10 M/21 F | 13 M/19 F | 17 M / 26 F | 0.39b |
| Number of episodes | 1.93±2.19 | 2.16±1.19 | N/A | 0.35c |
| Age of illness onset (years) | 26.41±10.72 | 24.58±10.42 | N/A | 0.26c |
| Duration of illness (months) | 40.68±50.41 | 39.14±56.62 | N/A | 0.46c |
| HAMD-24 | 26.76±5.58 | 26.23±6.24 | N/A | 0.37c |
| YRMS | 2.31±3.69 | 1.58±1.50 | N/A | 0.16c |
| Mean FD (mm) | 0.09±0.06 | 0.09± 0.04 | 0.08±0.03 | 0.42a |
Abbreviations: ANOVA, analysis of variance; BD, bipolar disorder; FD, framewise displacement; HAMD, Hamilton Depression Scale; HC, healthy control; N/A, not applicable; UD, unipolar depression; YRMS, Young Mania Rating Scale.
The P-value was obtained from a permutation ANOVA analysis.
The P-value was obtained from Pearson’s χ2-test.
The P-value was calculated from a permutation two-sample t-test.
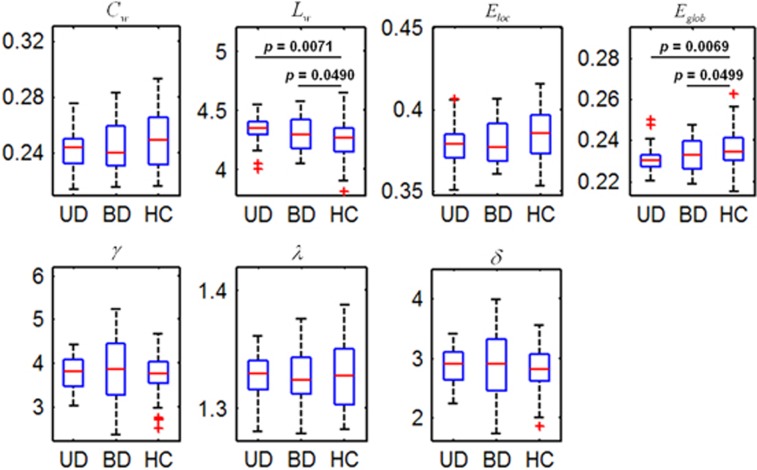
Global parameters
Figure 1 shows the global parameters (Cw, Lw, Eloc, Eglob, γ, λ and δ) for the UD, BD and control groups. Statistical analyses revealed significant differences in Lw and Eglob between the three groups (P<0.05). Post hoc comparisons revealed that both the UD and BD II groups had a significantly increased Lw and a significantly decreased Eglob, compared with the controls (P<0.05). However, no differences were found between the UD and BD II groups in these parameters (see Supplementary Table S2 for more information).
Figure 1.
Box plots showing statistical comparisons in the global parameters between the patients with UD, patients with BDs and HCs. The symbol of '+' in red color indicates outliers. Significant group effects were observed in Lw (P=0.0335) and Eglob (P=0.028). δ, small worldness; γ, normalized clustering coefficient; λ, normalized shortest path length; BD, bipolar disorder; Cw, weighted clustering coefficient; Eglob, weighted global efficiency; Eloc, weighted local efficiency; HC, healthy control; Lw, weighted characteristic path length; UD, unipolar depression.
Modularity
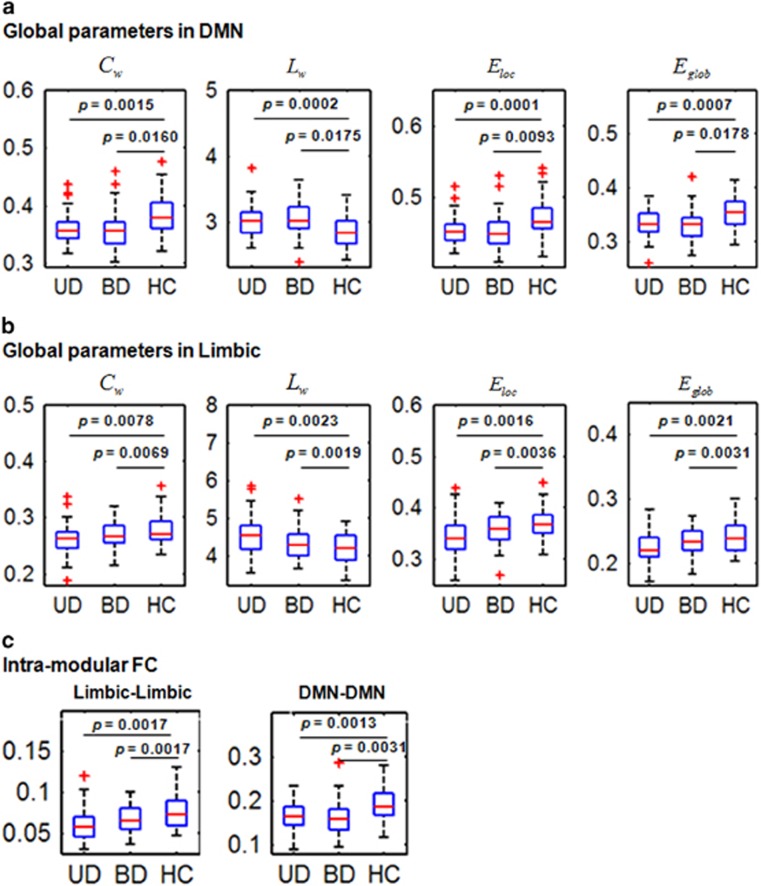
In the controls, we identified seven modules (Q=0.6803): the sensorimotor network, cerebellum, visual network, limbic network, frontoparietal network, DMN and salience network, which are shown in Supplementary Figure S3. The limbic network primarily includes the bilateral hippocampus, parahippocampus, thalamus, anterior cingulate gyrus, temporal pole, caudate and amygdala. The DMN primarily involves the bilateral medial frontal gyrus, superior frontal gyrus, precuneus, posterior cingulate gyrus, angular gyrus, middle temporal gyrus and inferior temporal gyrus. The sensorimotor network includes the bilateral precentral gyrus and postcentral gyrus. The visual network includes the bilateral occipital gyrus. The frontoparietal network involves the bilateral inferior parietal gyrus, middle frontal gyrus, inferior frontal gyrus, superior frontal gyrus and middle temporal gyrus. The salience network involves the bilateral insular and cingulate gyrus. These were consistent with previous studies.54, 55
We calculated four global parameters (Cw, Lw, Eloc and Eglob) for each module as well as the intra- and intermodular FCs for each subject. The statistical analysis for these seven modules, except for the DMN and the limbic network, showed no significant group effect for any of these four parameters. Figure 2a shows that for the DMN, the four global parameters were all significantly different between the three groups (P<0.05, FDR-corrected; Supplementary Table S2). Post hoc comparisons revealed that both the UD and BD II groups had a significantly decreased Cw, Eloc and Eglob, but significantly increased Lw, compared with the controls. Figure 2b shows that, for the limbic network, the four global parameters were significantly different between the three groups (P<0.05, FDR-corrected; Supplementary Table S2). Post hoc comparisons found that both the UD and BD II groups had a significantly decreased Cw, Eloc and Eglob, but a significantly increased Lw, compared with the controls. However, no differences in these parameters were found between the UD and BD II groups.
Figure 2.
Box plots showing statistical comparisons in the global parameters and intramodular FC between the patients with UD, patients with BDs and HCs (permutation ANOVA, P<0.05, corrected) in the modules of the DMN and the limbic network. The symbol of '+' in red color indicates outliers. (a) For the DMN, we observed significant group effects in Cw (P=0.0147), Lw (P=0.0037), Eloc (P=0.0045) and Eglob (P=0.0026). (b) For the limbic network, we observed significant group effects in Cw (P=0.0068), Lw (P=0.0008), Eloc (P=0.0011) and Eglob (P=0.0019). (c) For the intramodular FC, we observed significant group effects in the DMN (P=0.0020) and the limbic network (P=0.0064). δ, small worldness; γ, normalized clustering coefficient; λ, normalized shortest path length; ANOVA, analysis of variance; BD, bipolar disorder; Cw, weighted clustering coefficient; DMN, default mode network; Eglob, weighted global efficiency; Eloc, weighted local efficiency; FC, functional connectivity; HC, healthy control; Lw, weighted characteristic path length; UD, unipolar depression.
Figure 2c shows that, of the seven modules, the intramodular FCs in the DMN and in the limbic network were significantly different between the three groups (P<0.05, FDR-corrected; Supplementary Table S3). Post hoc comparisons revealed that the UD and BD II groups had significantly decreased intramodular FCs in both the DMN and the limbic network compared with the controls. However, no differences in the intramodular FC in the DMN and the limbic network were found between the UD and BD II groups. For the other modules, we found no significant group effect in the intramodular FC.
The seven modules gave 28 intermodular FCs (
intermodular FCs). However, no significant group effect survived after FDR correction (P<0.05, FDR-corrected).
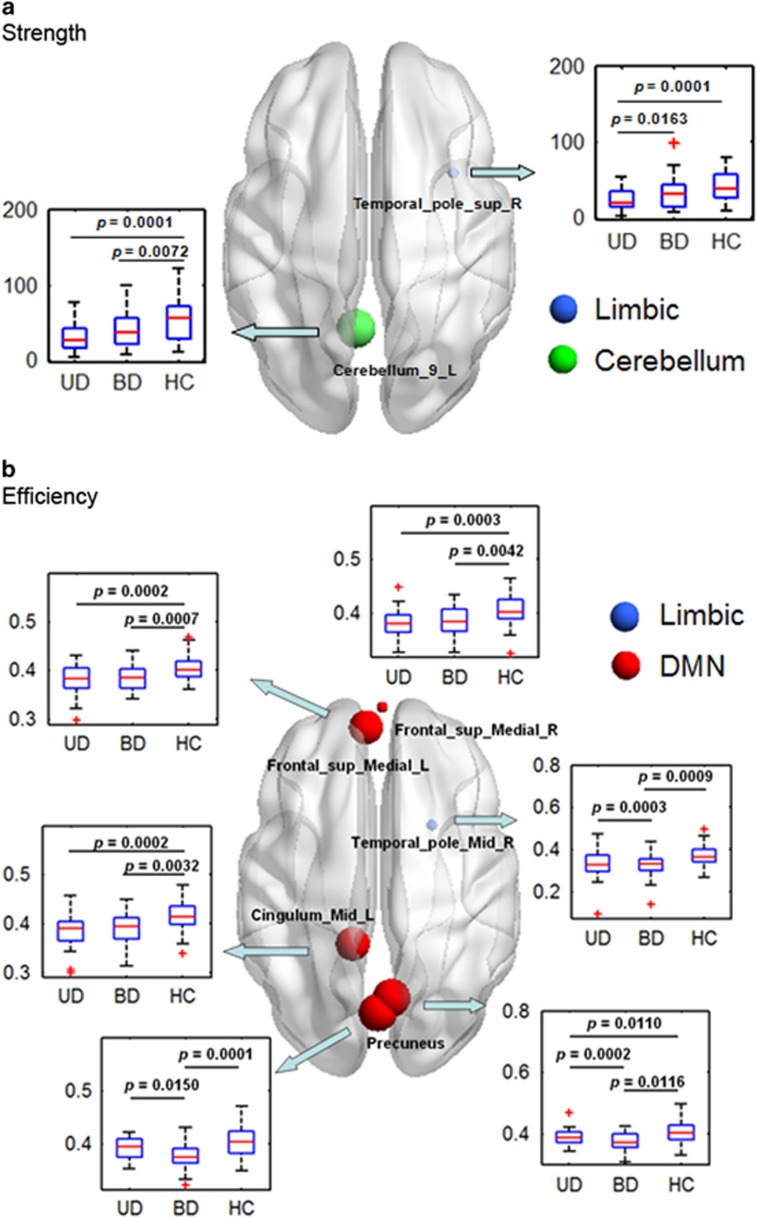
Nodal parameters
Figure 3 shows the brain regions with a significant group effect in the nodal parameters (
and
) between the three groups (P<0.05, FDR-corrected). We found a significant group effect in
in two regions (Figure 3a), which are located in the right superior temporal pole and left posterior lobe of the cerebellum. Post hoc comparisons revealed that, in the right superior temporal pole, the UD group had a significantly decreased
compared with the BD II group and with the control group. In the left cerebellum posterior lobe, both the UD and BD II groups had a significantly decreased
compared with the controls. In addition, we detected a significant group effect in
in six regions (Figure 3b), which are located in the bilateral medial superior frontal gyri, bilateral precuneus, right middle temporal pole and left middle cingulum. Post hoc comparisons revealed that in the right precuneus both the UD and BD II groups had a significantly decreased
compared with the controls and the UD group had a significantly increased
compared with the BD II group. In the bilateral medial superior frontal gyri and left middle cingulum, both the UD and BD II groups had a significantly decreased
compared with the control group. In addition, in the right middle temporal pole and left precuneus, the UD group showed a significantly increased
compared with the BD II and control groups. Furthermore, we noticed that most of these regions (1/2 for the
and 6/6 for the
) are included in the DMN and the limbic network (see Supplementary Table S4 for more information).
Figure 3.
Rendered plots of the brain regions showing significant group effects in nodal parameters (P<0.05, FDR-corrected). (a) Nodal strength; (b) nodal efficiency. Box plots showing statistical comparisons in the nodal parameters between the patients with UD, patients with BD and HC. The size of a node is inversely proportional to the P-value of the group effect for the given nodal parameter. The symbol of '+' in red color indicates outliers. BD, bipolar disorder; DMN, default mode network; FDR, false discovery rate; HC, healthy control; UD, unipolar depression.
Relationship between network parameters and clinical variables
For the correlations between network parameters and clinical variables, no significant correlation results survived after FDR correction (P<0.05, FDR-corrected). We have also displayed the correlation results without correction in Supplementary Figure S5.
Robustness
Table 2 lists the statistical results between the UD, BD II and control groups using four other data-processing strategies. These calculations indicated that most of our findings reported above were relatively reliable across the various strategies. Specifically, significant differences were always found in the the Lw and Eglob between the UD, BD II and control groups.
Table 2. Robustness of the statistical comparisons of the global parameters for the brain functional networks obtained with different strategies between the UD, BDs and control groups (P<0.05).
| Different processing strategies | Cw | Lw | Eloc | Eglob | δ |
|---|---|---|---|---|---|
| Global regressing | |||||
| Main processing strategy | – | S | – | S | >1 |
| Sparsity | – | S | – | S | >1 |
| No global regressing | – | S | – | – | >1 |
| Both positive and negative | – | S | – | S | >1 |
| Binary | – | S | – | S | >1 |
| No global regressing | |||||
| Positive | – | S | – | – | >1 |
| Both positive and negative | S | S | – | S | >1 |
Abbreviations: δ, small worldness; BD, bipolar disorder; Cw, weighted clustering coefficient; Eglob, weighted global efficiency; Eloc, weighted local efficiency; FC, functional connectivity; Lw, weighted characteristic path length; R-fMRI, resting-state functional magnetic resonance imaging; UD, unipolar depression. Bold values are significant at P<0.05.
Notes: The ‘S’ indicates a significant group effect. ‘–’ indicates no significant group effect. ‘Sparsity’ stands for estimating global parameters in the threshold range of 0.10–0.35 at an interval of 0.01 by using a measure of sparsity (the ratio between the total number of edges and the maximum possible number of edges in a network). We applied these thresholds to each of the FC matrices. ‘No global regressing’ refers to a network analysis based on a connectivity matrix that was constructed from the R-fMRI data without regressing out the global signal. ‘Both positive and negative’ refers to a network analysis based on a connectivity matrix that included both positive and negative internodal correlations. ‘Positive’ refers to a network analysis based on a connectivity matrix that only included positive internodal correlations. In addition, ‘Binary’ refers to a connectivity matrix that was converted into a binary matrix at a selected threshold when constructing the network.
Bold values are significant at P<0.05.
Classification
For the network parameters showing significant group effects, we plotted their ROC curves to examine their ability to distinguish the UD patients from the controls, the BD II patients from the controls and the BD II patients from the UD patients. We found that the
in the right superior temporal pole provided the highest classification power for discriminating the UD patients from the controls (AUC=0.76, P=7.4e−6, 95% confidence interval (CI) area=0.64–0.87, sensitivity=61.29%, specificity=81.40%, efficiency=72.97% Supplementary Table S5) and the
in the right precuneus provided the highest classification power for discriminating the BD II patients from the controls (AUC=0.75, P=6.4e−6, 95% CI area=0.64–0.87, sensitivity=78.13%, specificity=62.79%, efficiency=69.33% Supplementary Table S6). In addition, we found that the
in the left precuneus provided the highest discrimination ability for distinguishing the BD II patients from the UD patients (AUC=0.65, P=1.7e−2, 95% CI area=0.51–0.78, sensitivity=37.50%, specificity=93.55%, efficiency=65.08% Supplementary Table S7).
Discussion
By studying these unmedicated UD patients and unmedicated BD II-depressed patients, we detected abnormal topological parameters of the brain functional networks without the potentially confounding influence of psychotropic medications such as lithium or antidepressants. The main findings are as follows: (1) at the global level, both the UD and the BD II patients showed a significantly increased Lw and a decreased Eglob; (2) at the module level, both the UD and BD II patients had disrupted intramodular connectivity within the DMN and limbic network; (3) at the nodal level, both the UD and BD II patients showed impaired nodal parameters predominantly in regions of the DMN, limbic network and cerebellum; (4) the UD and BD II groups showed differences in the nodal parameters in the precuneus and temporal pole. To our knowledge, this study is the first to report topologically convergent and divergent patterns of the whole-brain functional networks between UD and BD II patients. These findings may contribute to our understanding of the neuropathological mechanisms in these two disorders.
Similar global-parameter changes in UD and BD
In this study, the whole-brain functional networks in the UD, BD II and control groups showed small-world properties that were consistent with previous findings in healthy adults.22, 56 We also found several global topological properties of the whole-brain networks, including increased characteristic path length Lw and decreased global efficiency Eglob (Figure 1), that showed the same change in direction in the UD and BD II groups compared with the controls. These findings, which were consistent with previous neuroimaging studies of brain functional and structural networks in UD patients28, 57, 58 and BD patients,32, 59, 60 may reflect the disrupted global integration of the whole-brain functional networks in the two disorders. We also noticed inconsistency between this study and several previous studies in the changes in the global properties in the UD patients. For example, Zhang et al.31 found decreased characteristic path length Lw and increased global efficiency Eglob in first-episode drug-naive UD patients by measuring the partial correlation coefficients of the R-fMRI signals between 90 cortical and subcortical regions; this is the opposite of what we found. In addition, several R-fMRI studies found no differences in global topological properties between UD patients and healthy controls.61, 62, 63 These inconsistencies may have been caused by any of several factors, including sample heterogeneity (age, depressive episode, medication and disease duration), modality used (for example, EEG/MEG; structural, diffusion and functional MRI), selection of the network matrix (binary or weighted) or the definitions of node and edge weight.
Similar modular-parameter changes in UD and BD
In this study, both the UD and BD II groups had decreased clustering coefficients Cw, local efficiencies Eloc, global efficiencies Eglob and increased characteristic path lengths Lw within two modules: the DMN and the limbic network (Figures 2a and b). This may indicate disrupted effective segregation and integration of information processing within these networks. In addition, the UD and BD II groups had decreased intramodular FCs in the DMN and limbic network (Figure 2c). These findings suggest that UD and BD patients share similar disruptions of their intrinsic modular structures. In fact, previous studies have suggested that the DMN is involved in a diverse array of functions, such as episodic memory, self-relevant mental processing, monitoring the external environment, remembering the past and planning the future.64, 65 Several studies have also found DMN abnormality in UD5, 31, 64, 66 and BD8, 67 patients. Aberrant DMN function could lead to dysfunctional self-referential and affective processing in the form of an excessively negative self-focus.68, 69 The limbic system is the central part of the ‘emotional brain’ circuitry and is dedicated to processing and regulating emotion or participating in emotional expression.70, 71 Previous studies have reported brain structural and functional abnormalities in the limbic system supporting emotion dysregulation in UD and BD patients,4, 72 and several studies found dysfunctional limbic circuits in depression with clinical remission, regardless of the specific treatment modality.72, 73 Taken together, our findings of disruptions in the DMN and the limbic network in UD and BD II may contribute to core deficits in cognitive and affective functioning that are believed to underlie clinical symptoms.
Similar and distinct nodal parameter changes in UD and BD
In addition to the global and modular levels, we found that both the UD and BD II groups showed decreased nodal strength and nodal efficiency predominantly in the DMN and the limbic network, including the bilateral precuneus, left middle cingulum, bilateral superior frontal gyri and right temporal pole (Figure 3 and Supplementary Table S4), which suggests impaired regional connectivity in these regions. These results are consistent with several previous studies related to UD and BD, which reported that these regions exhibited abnormalities in FC and local activation,5, 6, 8, 74 glucose metabolism75 and gray matter morphology.6 In addition, we found decreased nodal strength in the left posterior lobe of the cerebellum in both the UD and BD II groups compared with the controls (Figure 3a). In fact, other studies have shown cerebellum abnormalities, including reduced gray matter volume,76, 77, 78 decreased spontaneous activity18, 78, 79 and decreased FC,18 in both UD and BD patients. A recent R-fMRI study also found increased cerebellar–DMN connectivity in depressed patients compared with healthy controls.80 Therefore, our finding of impaired connectivity in the posterior lobe of the cerebellum provides additional evidence for the involvement of cerebellar dysfunction in the pathophysiology of UD and BD.
We also found significant differences in nodal parameters (nodal strength and nodal efficiency) between the UD and BD II patients in the bilateral precuneus and right temporal pole (Figure 3), suggesting pathophysiological differences between UD and BD. Specifically, BD patients may display more significant abnormalities than UD patients in these regions. A further ROC analysis showed that abnormal nodal strength in the right superior temporal pole was specific to the UD group (Supplementary Table S5), whereas abnormal nodal efficiency in the right precuneus was specific to the BD II group (Supplementary Table S6). Nevertheless, a larger sample size is warranted in future studies to replicate our results and to extend the generalizability of our results. We also found that nodal efficiency in the precuneus had high classification power in discriminating the BD group from the UD group (Supplementary Table S7). These results are partially consistent with previous studies. A R-fMRI study found that the increase in the intrinsic connectivity of the amygdala with the temporal poles was inversely correlated with depression severity in UD.81 Task-based fMRI studies found increased amygdala activity in response to angry82 and sad12, 83 faces, but decreased amygdala activity in response to positive12, 83 faces in UD compared with participants with BD. These changes in the amygdala may represent a potential neural basis for the increased negative and reduced positive emotional attentional bias in UD. In addition, a structural MRI study found a gray matter volume difference between UD and BD in the amygdala.77 Thus, these findings suggest that abnormalities in the temporal pole and amygdala supporting emotion processing may be specific to UD, although the underlying mechanisms remain to be clarified.77, 84 In addition, Marchand et al.16 suggested that the FC of the right posterior cingulate cortex can be used to differentiate bipolar type II and UD. Recently, a voxel-based meta-analysis of diffusion tensor imaging studies revealed a greater decrease in fractional anisotropy in the left posterior cingulum in BD compared with UD.85 A number of studies support the importance of the posterior cingulum and precuneus/posterior cingulate cortex in neurocognitive functions such as memory, attention and planning.86 Therefore, these studies, combined with our findings of reduced nodal efficiency in the precuneus in BD, suggest that cognitive deficits may be more representative of BD,85, 87 although a degree of cognitive decline has been described in both UD and BD.88 Finally, our results of differences in nodal parameters in the precuneus and temporal pole may aid in distinguishing the two types of depression.
Limitations
This study has several limitations. First, we cannot predict whether some patients with depression diagnosed as UD will later switch to BD, although the patients with UD in this study had no family history of BD. In addition, we tracked the UD patients after we acquired the data and found that no UD patient had switched to BD by the time of the submission of this manuscript. Second, this study did not consider the effect of brain structural changes on the network topologies in the UD or BD patients, which may bias the findings of the present study. Several previous studies suggested that brain structural changes may have an impact on the results of FC and network analysis.28 Third, we only examined patients with BD II, thereby avoiding the potential confounds of the bipolarity subtype differences in brain functional networks but also limiting the generalizability of our findings to BD I. Fourth, although the participants were unmedicated for at least 5 months prior to the scan, each group may have had variable levels of past exposure. Thus, it is difficult to estimate the potential contribution of lifetime exposure to medication in relation to the results. Fifth, because clinical symptomatology could change over a period of 7 days, collecting the measurements of clinical variables such as the HAMD and Young Mania Rating Scale scores over a 7-day period is another limitation. Sixth, because of selection bias,89 the ROC results should be interpreted as descriptive and exploratory, requiring confirmation in an independent data set. Further studies utilizing optimized analytical strategies and multiple imaging modalities are needed to detect more sensitive and robust biomarkers for UD and BD. Finally, like all R-fMRI studies, our results are based on the FC analysis, which is only an exploratory technique and requires confirmation with task-based or experimental work.
In conclusion, our study revealed convergent deficits in the topological organization in the whole brain and in the DMN and the limbic network in UD and BD II. These deficits may reflect overlapping pathophysiological processes in unipolar and bipolar depression. In addition, divergent regional connectivity supporting emotion processing could potentially provide biomarkers that would aid in differentiating between these disorders.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81671670, 81501456, 81471650, 81371535, 81428013 and 81471654); Planned Science and Technology Project of Guangdong Province, China (2014B020212022); and Planned Science and Technology Project of Guangzhou, China (1563000653, 155700029, 20160402007 and 201604020184). The funding organizations had no further role in the study design, data collection, analysis and interpretation, or the paper writing. We express our appreciation to Drs Rhoda E and Edmund F Perozzi for editing assistance, and thank the four anonymous reviewers for their constructive comments and their suggestions.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
The authors declare no conflict of interest.
Supplementary Material
References
- Fountoulakis KN. The emerging modern face of mood disorders: a didactic editorial with a detailed presentation of data and definitions. Ann Gen Psychiatry 2010; 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet 2016; 387: 1561–1572. [DOI] [PubMed] [Google Scholar]
- Cardoso de Almeida JR, Phillips ML. Distinguishing between unipolar depression and bipolar depression: current and future clinical and neuroimaging perspectives. Biol Psychiatry 2013; 73: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio G, Fossati P, Boyer P, Brambilla P, Falkai P, Gruber O et al. Common and distinct neural correlates of emotional processing in bipolar disorder and major depressive disorder: a voxel-based meta-analysis of functional magnetic resonance imaging studies. Eur Neuropsychopharmacol 2012; 22: 100–113. [DOI] [PubMed] [Google Scholar]
- Gong Q, He Y. Depression, neuroimaging and connectomics: a selective overview. Biol Psychiatry 2015; 77: 223–235. [DOI] [PubMed] [Google Scholar]
- Kempton MJ, Salvador Z, Munafo MR, Geddes JR, Simmons A, Frangou S et al. Structural neuroimaging studies in major depressive disorder. Meta-analysis and comparison with bipolar disorder. Arch Gen Psychiatry 2011; 68: 675–690. [DOI] [PubMed] [Google Scholar]
- Lui S, Zhou XJ, Sweeney JA, Gong Q. Psychoradiology: the frontier of neuroimaging in psychiatry. Radiology 2016; 281: 357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas C, Lopez-Jaramillo C, Vieta E. A systematic literature review of resting state network—functional MRI in bipolar disorder. J Affect Disord 2013; 150: 727–735. [DOI] [PubMed] [Google Scholar]
- Serafini G, Pompili M, Borgwardt S, Houenou J, Geoffroy PA, Jardri R et al. Brain changes in early-onset bipolar and unipolar depressive disorders: a systematic review in children and adolescents. Eur Child Adolesc Psychiatry 2014; 23: 1023–1041. [DOI] [PubMed] [Google Scholar]
- Silverstone T, McPherson H, Li Q, Doyle T. Deep white matter hyperintensities in patients with bipolar depression, unipolar depression and age-matched control subjects. Bipolar Disord 2003; 5: 53–57. [DOI] [PubMed] [Google Scholar]
- Versace A, Almeida JR, Quevedo K, Thompson WK, Terwilliger RA, Hassel S et al. Right orbitofrontal corticolimbic and left corticocortical white matter connectivity differentiate bipolar and unipolar depression. Biol Psychiatry 2010; 68: 560–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotegerd D, Stuhrmann A, Kugel H, Schmidt S, Redlich R, Zwanzger P et al. Amygdala excitability to subliminally presented emotional faces distinguishes unipolar and bipolar depression: an fMRI and pattern classification study. Hum Brain Mapp 2014; 35: 2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Kable JW, Vandekar L, Katchmar N, Bassett DS, Baldassano CF et al. Common and dissociable dysfunction of the reward system in bipolar and unipolar depression. Neuropsychopharmacology 2015; 40: 2258–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goya-Maldonado R, Brodmann K, Keil M, Trost S, Dechent P, Gruber O. Differentiating unipolar and bipolar depression by alterations in large-scale brain networks. Hum Brain Mapp 2016; 37: 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu X, Zhang J, Guo X, Long Z, Yao L. Altered effective connectivity model in the default mode network between bipolar and unipolar depression based on resting-state fMRI. J Affect Disord 2015; 182: 8–17. [DOI] [PubMed] [Google Scholar]
- Marchand WR, Lee JN, Johnson S, Gale P, Thatcher J. Differences in functional connectivity in major depression versus bipolar II depression. J Affect Disord 2013; 150: 527–532. [DOI] [PubMed] [Google Scholar]
- Anand A, Li Y, Wang Y, Lowe MJ, Dzemidzic M. Resting state corticolimbic connectivity abnormalities in unmedicated bipolar disorder and unipolar depression. Psychiatry Res 2009; 171: 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhong S, Jia Y, Zhou Z, Wang B, Pan J et al. Interhemispheric resting state functional connectivity abnormalities in unipolar depression and bipolar depression. Bipolar Disord 2015; 17: 486–495. [DOI] [PubMed] [Google Scholar]
- Abe C, Ekman CJ, Sellgren C, Petrovic P, Ingvar M, Landen M. Cortical thickness, volume and surface area in patients with bipolar disorder types I and II. J Psychiatry Neurosci 2016; 41: 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caseras X, Lawrence NS, Murphy K, Wise RG, Phillips ML. Ventral striatum activity in response to reward: differences between bipolar I and II disorders. Am J Psychiatry 2013; 170: 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CT, Hsieh JC, Wang SJ, Yang BH, Bai YM, Lin WC et al. Differential relations between fronto-limbic metabolism and executive function in patients with remitted bipolar I and bipolar II disorder. Bipolar Disord 2012; 14: 831–842. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 2009; 10: 186–198. [DOI] [PubMed] [Google Scholar]
- Singh MK, Kesler SR, Hadi Hosseini SM, Kelley RG, Amatya D, Hamilton JP et al. Anomalous gray matter structural networks in major depressive disorder. Biol Psychiatry 2013; 74: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X, He Y, Yu X, Wang H. Apolipoprotein E epsilon4 modulates functional brain connectome in Alzheimer's disease. Hum Brain Mapp 2015; 36: 1828–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos Siqueira A, Biazoli Junior CE, Comfort WE, Rohde LA, Sato JR. Abnormal functional resting-state networks in ADHD: graph theory and pattern recognition analysis of fMRI data. Biomed Res Int 2014; 2014: 380531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lutterveld R, Diederen KM, Otte WM, Sommer IE. Network analysis of auditory hallucinations in nonpsychotic individuals. Hum Brain Mapp 2014; 35: 1436–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Gao C, Chen C, Ma S, Netra R, Wang Y et al. A preliminary study of the dysregulation of the resting networks in first-episode medication-naive adolescent depression. Neurosci Lett 2011; 503: 105–109. [DOI] [PubMed] [Google Scholar]
- Meng C, Brandl F, Tahmasian M, Shao J, Manoliu A, Scherr M et al. Aberrant topology of striatum's connectivity is associated with the number of episodes in depression. Brain 2014; 137(Pt 2): 598–609. [DOI] [PubMed] [Google Scholar]
- Tao H, Guo S, Ge T, Kendrick KM, Xue Z, Liu Z et al. Depression uncouples brain hate circuit. Mol Psychiatry 2013; 18: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dai Z, Peng H, Tan L, Ding Y, He Z et al. Overlapping and segregated resting-state functional connectivity in patients with major depressive disorder with and without childhood neglect. Hum Brain Mapp 2014; 35: 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang J, Wu Q, Kuang W, Huang X, He Y et al. Disrupted brain connectivity networks in drug-naive, first-episode major depressive disorder. Biol Psychiatry 2011; 70: 334–342. [DOI] [PubMed] [Google Scholar]
- Spielberg JM, Beall EB, Hulvershorn LA, Altinay M, Karne H, Anand A. Resting state brain network disturbances related to hypomania and depression in medication-free bipolar disorder. Neuropsychopharmacology 2016; 41: 3016–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao-Gan Y, Yu-Feng Z. DPARSF: A MATLAB toolbox for "pipeline" data analysis of resting-state fMRI. Front Syst Neurosci 2010; 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X, Xia M, Liao X, Evans A, He Y. GRETNA: a graph theoretical network analysis toolbox for imaging connectomics. Front Hum Neurosci 2015; 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15: 273–289. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. Neuroimage 2010; 53: 1197–1207. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R. Movement-related effects in fMRI time-series. Magn Reson Med 1996; 35: 346–355. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu M, Fan Y, Wen X, Zhang R, Wang B et al. Exploring brain functional plasticity in world class gymnasts: a network analysis. Brain Struct Funct 2016; 221: 3503–3519. [DOI] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J Neurophysiol 2009; 101: 3270–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage 2009; 44: 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C. Correlations and anticorrelations in resting-state functional connectivity MRI: a quantitative comparison of preprocessing strategies. Neuroimage 2009; 47: 1408–1416. [DOI] [PubMed] [Google Scholar]
- Wang JH, Zuo XN, Gohel S, Milham MP, Biswal BB, He Y. Graph theoretical analysis of functional brain networks: test-retest evaluation on short- and long-term resting-state functional MRI data. PLoS ONE 2011; 6: e21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage 2010; 52: 1059–1069. [DOI] [PubMed] [Google Scholar]
- Uehara T, Yamasaki T, Okamoto T, Koike T, Kan S, Miyauchi S et al. Efficiency of a "small-world" brain network depends on consciousness level: a resting-state FMRI study. Cereb Cortex 2014; 24: 1529–1539. [DOI] [PubMed] [Google Scholar]
- Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci USA 2006; 103: 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res 2003; 12: 419–446. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp 2002; 15: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001; 125: 279–284. [DOI] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral power analysis for the behavioral sciences. 2nd edn. L. Erlbaum Associates: Hillsdale, NJ, 1988, pp 20–26. [Google Scholar]
- Deuker L, Bullmore ET, Smith M, Christensen S, Nathan PJ, Rockstroh B et al. Reproducibility of graph metrics of human brain functional networks. Neuroimage 2009; 47: 1460–1468. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J et al. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data 2014; 1: 140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencina MJ, D'Agostino RB Sr., D'Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008; 27: 157–172, discussion 207-112. [DOI] [PubMed] [Google Scholar]
- Desco M, Hernandez JA, Santos A, Brammer M. Multiresolution analysis in fMRI: sensitivity and specificity in the detection of brain activation. Hum Brain Mapp 2001; 14: 16–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H et al. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PLoS ONE 2009; 4: e5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 2011; 106: 1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M, van den Heuvel MP, Fornito A, He Y, Hulshoff Pol HE, Agosta F et al. Assessment of system dysfunction in the brain through MRI-based connectomics. Lancet Neurol 2013; 12: 1189–1199. [DOI] [PubMed] [Google Scholar]
- Bai F, Shu N, Yuan Y, Shi Y, Yu H, Wu D et al. Topologically convergent and divergent structural connectivity patterns between patients with remitted geriatric depression and amnestic mild cognitive impairment. J Neurosci 2012; 32: 4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt V, Krause AL, Starck T, Nissila J, Timonen M, Kiviniemi V et al. Graph theory reveals hyper-functionality in visual cortices of seasonal affective disorder patients. World J Biol Psychiatry 2015; 16: 123–134. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Bolbecker AR, Howell J, Rass O, Sporns O, Hetrick WP et al. Disturbed resting state EEG synchronization in bipolar disorder: a graph-theoretic analysis. Neuroimage Clin 2013; 2: 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow A, Ajilore O, Zhan L, Arienzo D, GadElkarim J, Zhang A et al. Impaired inter-hemispheric integration in bipolar disorder revealed with brain network analyses. Biol Psychiatry 2013; 73: 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr IJ, Kenny E, Blamire A, O'Brien JT, Thomas AJ, Richardson J et al. Resting-state functional connectivity in late-life depression: higher global connectivity and more long distance connections. Front Psychiatry 2012; 3: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord A, Horn D, Breakspear M, Walter M. Changes in community structure of resting state functional connectivity in unipolar depression. PLoS ONE 2012; 7: e41282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Shi F, Shen T, Peng Z, Zhang C, Liu X et al. Altered brain network modules induce helplessness in major depressive disorder. J Affect Disord 2014; 168: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga-Barke EJ. Default-mode brain dysfunction in mental disorders: a systematic review. Neurosci Biobehav Rev 2009; 33: 279–296. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Menon V. Default-mode activity during a passive sensory task: uncoupled from deactivation but impacting activation. J Cogn Neurosci 2004; 16: 1484–1492. [DOI] [PubMed] [Google Scholar]
- Fang J, Rong P, Hong Y, Fan Y, Liu J, Wang H et al. Transcutaneous vagus nerve stimulation modulates default mode network in major depressive disorder. Biol Psychiatry 2015; 79: 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Ruano G, Windemuth A, O'Neil K, Berwise C, Dunn SM et al. Multivariate analysis reveals genetic associations of the resting default mode network in psychotic bipolar disorder and schizophrenia. Proc Natl Acad Sci USA 2014; 111: E2066–E2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TC, Connolly CG, Henje Blom E, LeWinn KZ, Strigo IA, Paulus MP et al. Emotion-dependent functional connectivity of the default mode network in adolescent depression. Biol Psychiatry 2015; 78: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S, Ford JM. Default mode network activity and connectivity in psychopathology. Annu Rev Clin Psychol 2012; 8: 49–76. [DOI] [PubMed] [Google Scholar]
- Dalgleish T. The emotional brain. Nat Rev Neurosci 2004; 5: 583–589. [DOI] [PubMed] [Google Scholar]
- Price JL, Drevets WC. Neurocircuitry of mood disorders. Neuropsychopharmacology 2010; 35: 192–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS. Limbic-cortical dysregulation: a proposed model of depression. J Neuropsychiatry Clin Neurosci 1997; 9: 471–481. [DOI] [PubMed] [Google Scholar]
- Mayberg HS. Modulating limbic-cortical circuits in depression: targets of antidepressant treatments. Semin Clin Neuropsychiatry 2002; 7: 255–268. [DOI] [PubMed] [Google Scholar]
- Stoddard J, Gotts SJ, Brotman MA, Lever S, Hsu D, Zarate C et al. Aberrant intrinsic functional connectivity within and between corticostriatal and temporal-parietal networks in adults and youth with bipolar disorder. Psychol Med 2016; 46: 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Momose T, Kasai K. Brain glucose metabolism difference between bipolar and unipolar mood disorders in depressed and euthymic states. Prog Neuropsychopharmacol Biol Psychiatry 2009; 33: 243–250. [DOI] [PubMed] [Google Scholar]
- Grieve SM, Korgaonkar MS, Koslow SH, Gordon E, Williams LM. Widespread reductions in gray matter volume in depression. Neuroimage Clin 2013; 3: 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlich R, Almeida JJ, Grotegerd D, Opel N, Kugel H, Heindel W et al. Brain morphometric biomarkers distinguishing unipolar and bipolar depression. A voxel-based morphometry-pattern classification approach. JAMA Psychiatry 2014; 71: 1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JR, Hewedi DH, Eissa AM, Moustafa AA. The cerebellum and psychiatric disorders. Front Public Health 2015; 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang MJ, Zhou Q, Yang KR, Yang XL, Fang J, Chen WL et al. Identify changes of brain regional homogeneity in bipolar disorder and unipolar depression using resting-state FMRI. PLoS ONE 2013; 8: e79999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Liu F, Liu J, Yu M, Zhang Z, Liu G et al. Increased cerebellar-default-mode-network connectivity in drug-naive major depressive disorder at rest. Medicine 2015; 94: e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubbu R, Konduru N, Cortese F, Bray S, Gaxiola-Valdez I, Goodyear B. Reduced intrinsic connectivity of amygdala in adults with major depressive disorder. Front Psychiatry 2014; 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier JC, Keener MT, Almeida J, Kronhaus DM, Phillips ML. Amygdala and whole-brain activity to emotional faces distinguishes major depressive disorder and bipolar disorder. Bipolar Disord 2013; 15: 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotegerd D, Suslow T, Bauer J, Ohrmann P, Arolt V, Stuhrmann A et al. Discriminating unipolar and bipolar depression by means of fMRI and pattern classification: a pilot study. Eur Arch Psychiatry Clin Neurosci 2013; 263: 119–131. [DOI] [PubMed] [Google Scholar]
- Almeida JR, Versace A, Mechelli A, Hassel S, Quevedo K, Kupfer DJ et al. Abnormal amygdala-prefrontal effective connectivity to happy faces differentiates bipolar from major depression. Biol Psychiatry 2009; 66: 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise T, Radua J, Nortje G, Cleare AJ, Young AH, Arnone D. Voxel-based meta-analytical evidence of structural disconnectivity in major depression and bipolar disorder. Biol Psychiatry 2016; 79: 293–302. [DOI] [PubMed] [Google Scholar]
- Delano-Wood L, Stricker NH, Sorg SF, Nation DA, Jak AJ, Woods SP et al. Posterior cingulum white matter disruption and its associations with verbal memory and stroke risk in mild cognitive impairment. J Alzheimers Dis 2012; 29: 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maalouf FT, Klein C, Clark L, Sahakian BJ, Labarbara EJ, Versace A et al. Impaired sustained attention and executive dysfunction: bipolar disorder versus depression-specific markers of affective disorders. Neuropsychologia 2010; 48: 1862–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel BD, Montali A, Gerra ML, Innamorati M, Girardi P, Pompili M et al. Cognitive impairment and its associations with the path of illness in affective disorders: a comparison between patients with bipolar and unipolar depression in remission. J Psychiatr Pract 2013; 19: 275–287. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Simmons WK, Bellgowan PS, Baker CI. Circular analysis in systems neuroscience: the dangers of double dipping. Nat Neurosci 2009; 12: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.