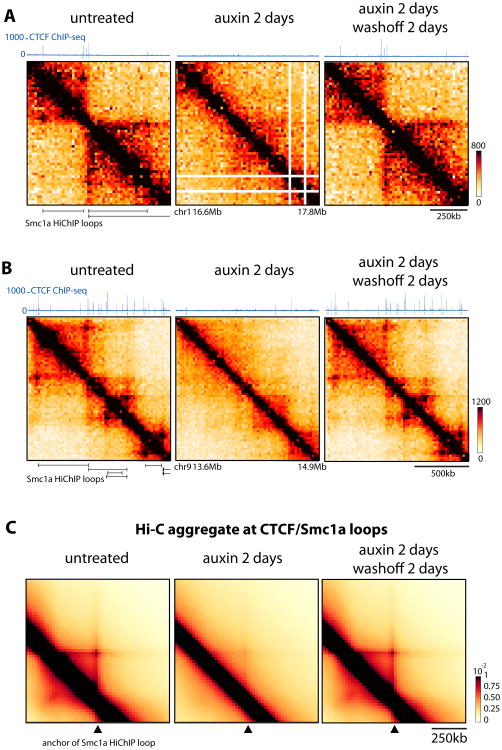
Figure 2. CTCF is required for accumulating loops between CTCF/Cohesin binding sites.
(A-B) Snapshots of 1.3 Mb of Hi-C data at 20kb resolution CTCF-AID mESCs aligned with CTCF ChIP-seq and the Smc1a HiChIP loops identified by Mumbach et al. 2016. Normalized Hi-C counts are multiplied by 105
(C) Genome-wide aggregation of normalized Hi-C signal anchored at Smc1a HiChip loops separated by 280 to 380kb (1196 loops). Similar results were obtained for smaller and larger loops. See Figure S2