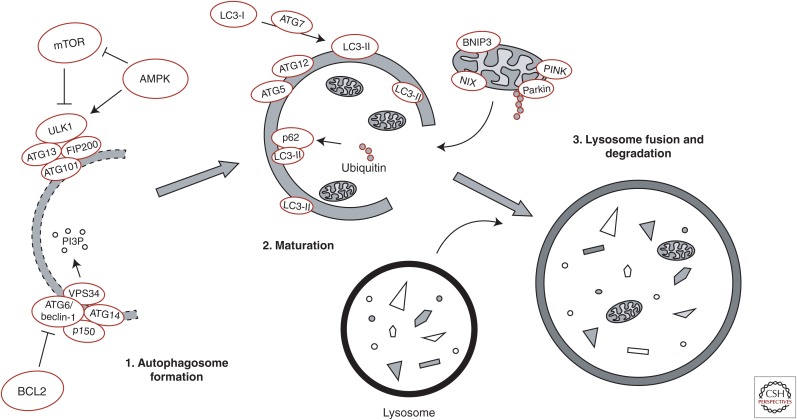
Figure 1.
Overview of basic molecular mechanisms involved in initiation of autophagy. The UNC-51-like kinase 1 (ULK1) kinase complex (including autophagy-related genes (ATG1)3, ATG101, and FAK family kinase-interacting protein of 200 kDa (FIP200)) and the class III PI3-kinase complex (including the catalytic vacuolar protein sorting mutant 34 (VPS34) and the regulatory ATG6/Beclin-1, p150, and ATG14) are stabilized on a phagophore membrane and generate a pool of phosphatidylinositol-3-phosphate (PI3P), which is essential for autophagosome formation. The 5′ AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) act as opposing regulators of ULK1 complex activity through stimulatory and inhibitory phosphorylation of ULK1, respectively, whereas AMPK inhibits mTOR activity. Disassociation of ATG6/Beclin-1 and B-cell lymphoma 2 (BCL2) is required for the assembly of the class III PI3-kinase complex. ATG7 regulates the recruitment of cytosolic LC3-I to the maturing autophagosome membrane, where ATG5-ATG12-dependent lipidation forms membrane-bound LC3-II. The autophagy adaptor protein p62 links ubiquitinated substrates with LC3-II. Selective autophagic clearance of mitochondria (mitophagy) is regulated by the kinase PTEN-induced putative kinase 1 (PINK1), which is stabilized on depolarized mitochondrial membranes and recruits the E3 ubiquitin ligase parkin to mitochondria. Parkin regulates ubiquitination of mitochondrial membrane proteins, thereby targeting mitochondria for degradation. The mitophagy receptors BCL2/adenovirus E1B 19-kDa protein-interacting protein 3 (BNIP3) and NIP3-like protein X (NIX) present on mitochondrial membranes can directly interact with LC3-II to induce mitophagy. Finally, the mature autophagosomes fuse with lysosomes causing hydrolytic degradation of autophagosomal cargo.