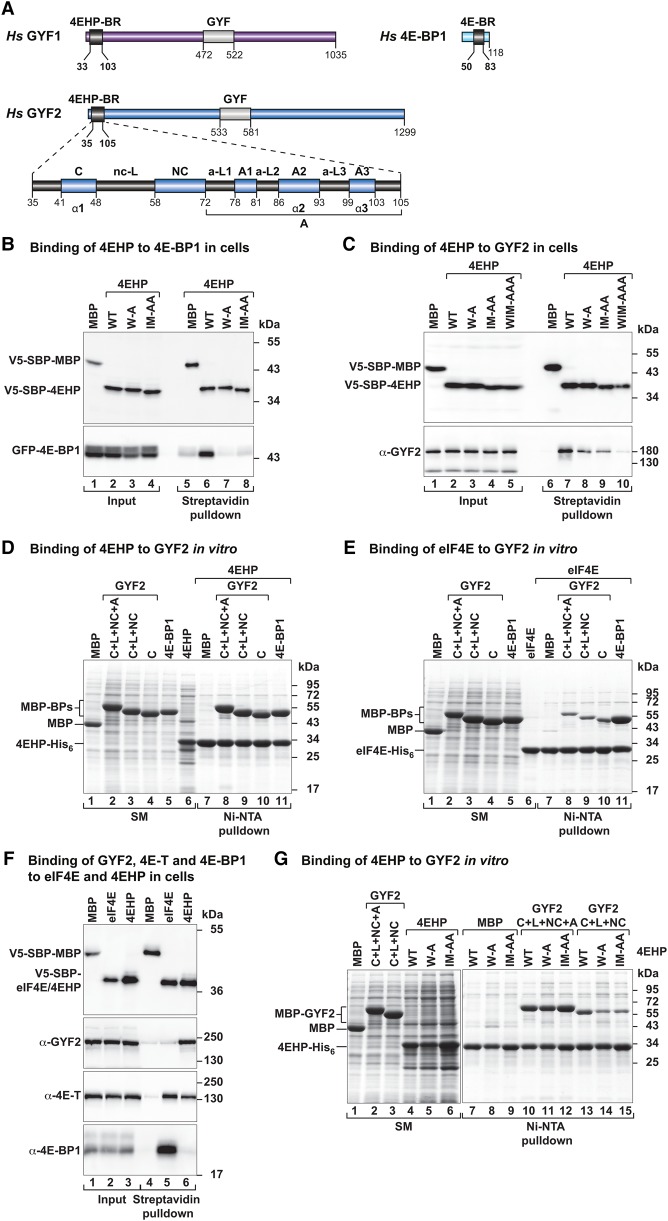
Figure 1.
GYF1/2 proteins use canonical, noncanonical, and auxiliary sequences to bind to 4EHP. (A) GYF1/2 proteins contain a central GYF domain and an N-terminal 4EHP-binding region (4EHP-BR). The 4EHP-BR includes canonical, noncanonical, and auxiliary motifs (A1–3) connected by linker sequences (nc-L and auxiliary linkers 1–3 [a-L1–3]). The 4E-binding region (4E-BR) of 4E-BP1 contains canonical and noncanonical motifs. (B,C) Western blots showing the interaction between V5-SBP-4EHP (wild type or the indicated mutants) and GFP-4E-BP1 (full-length) or endogenous GYF2. The proteins were pulled down using streptavidin-coated beads. V5-SBP-MBP (maltose-binding protein) served as negative control. The inputs (1.5% for the V5-tagged proteins and 1% for the GFP-tagged proteins) and bound fractions (3%–5% for the V5-tagged proteins and 20% for GYF2 and GFP-4E-BP1) were analyzed by Western blotting using anti-V5, anti-GFP, and anti-GYF2 antibodies. (D,E) Ni-NTA pull-down assays showing the interactions between GYF2 fragments (C+L+NC+A, C+L+NC, and C) and 4EHP-His6 (M1–F234) (D) or eIF4E-His6 (E). 4E-BP1 and MBP served as positive and negative controls, respectively. The GYF2 and 4E-BP1 peptides contain an N-terminal MBP tag and a C-terminal GB1 tag. The starting material (SM; 1.3% for MBPs and 6% for 4EHP and purified eIF4E) and bound fractions (7%–10%) were analyzed by SDS-PAGE followed by Coomassie blue staining. (F) The interaction between V5-SBP-eIF4E or 4EHP proteins and endogenous GYF2, 4E-T, and 4E-BP1 was analyzed in HEK293T cell lysates using streptavidin pull-downs. The input (1% for 4E-BP1 and 4E-T and 1.5% for V5-SBP-tagged proteins and GYF2) and bound fractions (20% for 4E-BP1 and 4ET, 30% for GYF2, and 5% for the V5-SBP-tagged proteins) were analyzed by Western blotting using the indicated antibodies. (G) Ni-NTA pull-down assay showing the interaction between 4EHP (M1–F234, wild type, or the indicated mutants) and GYF2 fragments. MBP served as a negative control. Samples were analyzed as described in D. The starting material (2% for the MBP-tagged proteins and 4%–12% for the 4EHP proteins) and bound fractions (10%) were analyzed by SDS-PAGE followed by Coomassie blue staining.