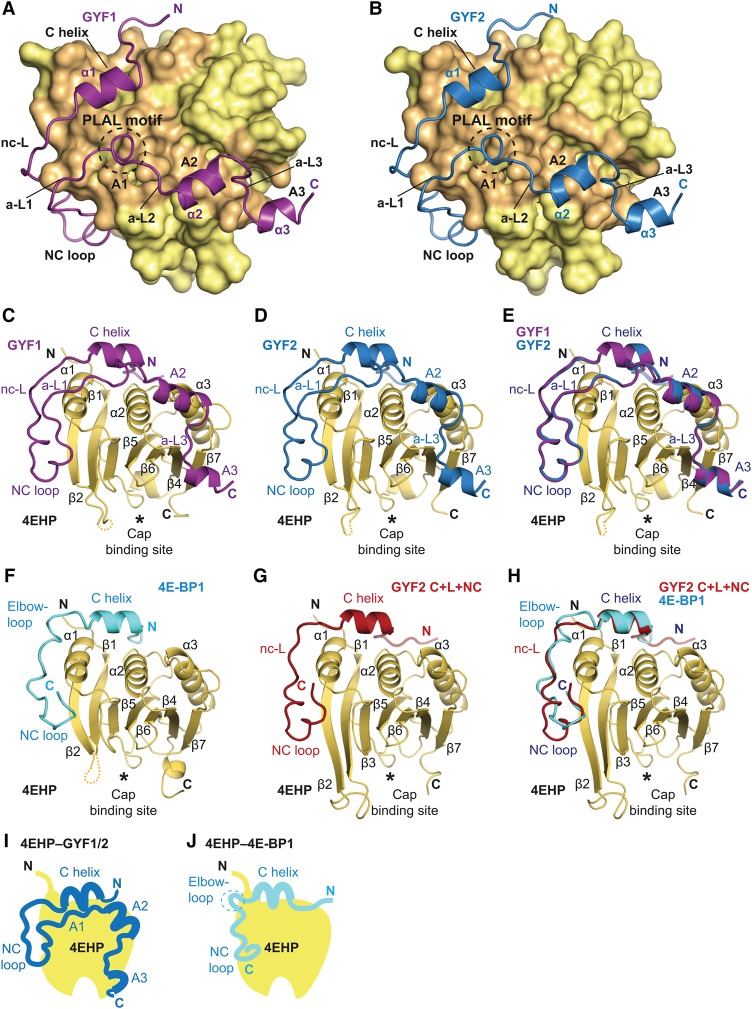
Figure 2.
Overall structures of GYF1, GYF2, and 4E-BP1 bound to 4EHP. (A,B) Overview of the structures of 4EHP bound to GYF1/2 (C+L+NC+A) fragments. The 4EHP surface is shown in yellow, and surface residues within a radius of 4 Å of the bound GYF1 or GYF2 peptides are colored in orange. The GYF1 and GYF2 peptides are colored in purple and blue, respectively. Selected secondary structure elements in the GYF1/2 peptides are indicated. The invariant PLAL motif of GYF1/2 is circled with a dashed line. (C,D) Cartoon representation of the structures of 4EHP bound to GYF1/2. Selected secondary structure elements are labeled in black for 4EHP and in color for GYF1/2. (E) Superposition of the structures of 4EHP bound to GYF1 and GYF2. For clarity, the 4EHP molecule from the 4EHP–GYF1 complex was omitted. The structures of the complexes are very similar, and overall root mean square deviations do not exceed 0.32 Å over 227 Cα atoms. (F) Structure of 4EHP bound to 4E-BP1. Selected secondary structure elements are labeled in black for 4EHP and in cyan for 4E-BP1. (G) Structure of 4EHP bound to the GYF2 C+L+NC fragment. Selected secondary structure elements are labeled in black and red for 4EHP and GYF2, respectively. (H) Superposition of the structures of 4EHP bound to the 4E-BP1 and GYF2 C+L+NC peptides. For clarity, the 4EHP molecule from the 4EHP–4E-BP1 complex was omitted. (I,J) Schematic representations of 4EHP bound to GYF1/2 and 4E-BP1.