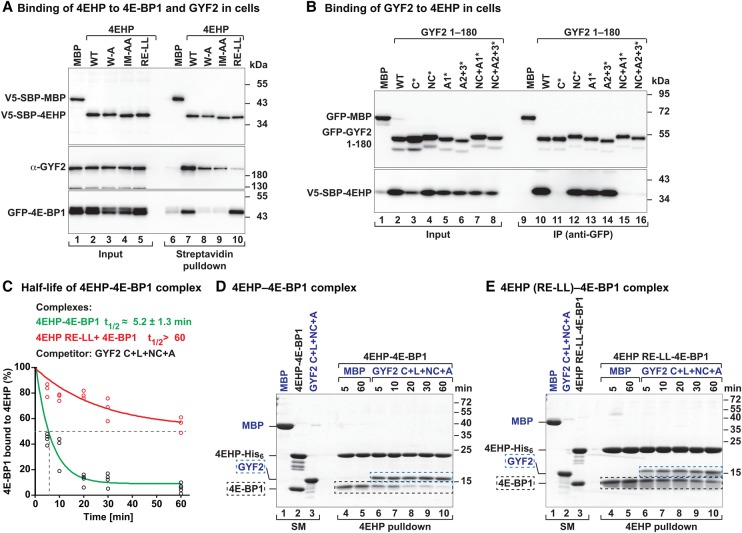
Figure 5.
The auxiliary interactions are crucial for the formation of the 4EHP–GYF complex. (A) Western blot showing the interaction of endogenous GYF2 or GFP-4E-BP1 with V5-SBP-4EHP (wild type or the indicated mutants). The proteins were pulled down using streptavidin-coated beads. The inputs (1.5% for the V5-tagged proteins and 1% for GYF2 and GFP-4E-BP1) and bound fractions (3% for the V5-tagged proteins and 20% for GYF2 and GFP-4E-BP1) were analyzed by Western blot using the indicated antibodies. (B) Interaction of V5-SBP-4EHP with GFP-GYF2 (residues 1–180; either wild type or the indicated mutants). The proteins were immunoprecipitated using anti-GFP antibodies. GFP-MBP served as negative control. The inputs (1.5% for the GFP-tagged proteins and 0.5% for V5-SBP-4EHP) and immunoprecipitates (7.5% for the GFP-tagged proteins and 30% for V5-SBP-4EHP) were analyzed by Western blot using anti-GFP and anti-V5 antibodies. (C–E) Purified 4EHP–4E-BP1 complexes containing 4EHP-His6 (wild type or the RE-LL mutant) were incubated in the presence of equimolar amounts of the GYF2 C+L+NC+A peptide C-terminally tagged with GB1 or MBP as a negative control. The proteins bound to 4EHP were pulled down using Ni-NTA beads at the indicated time points and analyzed by SDS-PAGE and Coomassie blue staining. C shows the quantification of the amount of 4E-BP1 still associated with 4EHP. n = 3. The half-life of the 4EHP–4E-BP1 complex (t1/2) in the presence of the competitor protein is represented as the mean ± SD. D and E show representative SDS-PAGE gels. The positions of the GYF2 and 4E-BP1 peptides are marked by blue and black dashed boxes, respectively. The lanes labeled SM (starting material) show the purified complexes and peptides used in the assay.