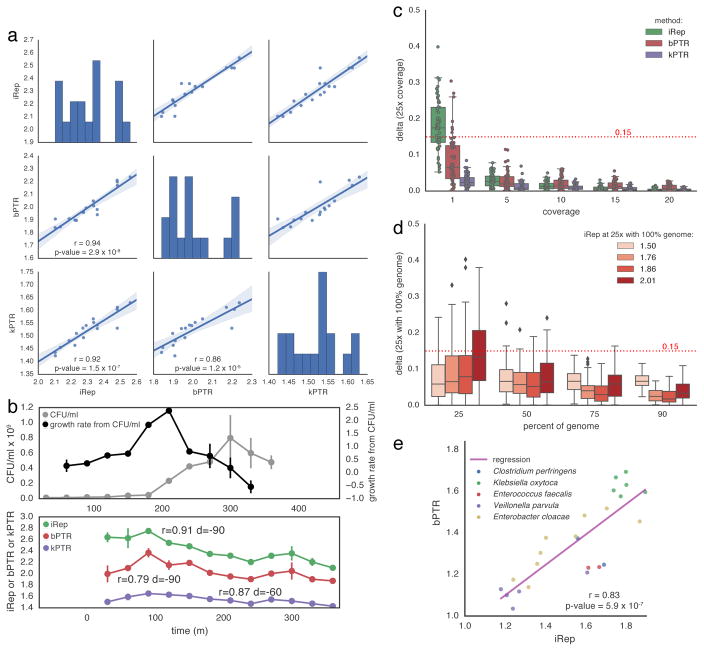
Figure 2. iRep is an accurate measure of in situ replication rates.
(a) iRep, bPTR, and kPTR measurements made for cultured Lactobacillus gasseri8 were compared (r = Pearson’s r value), showing strong agreement between all methods. (b) Colony forming unit (CFU) counts were available for a subset of these samples8, and used to calculate growth rates (n = 2). All methods were highly correlated with CFU-derived rates after first accounting for the delay between start of genome replication and observable change in population size (as noted previously8). Replication rates from CFU data were adjusted by variable amounts before calculating correlations with sequencing-based rates (best correlation shown; d = time adjustment). CFU data are plotted with a −90 minute offset. (c) Using the L. gasseri data, minimum coverage requirements were determined for each method by first measuring the replication rate at 25× coverage, and then comparing to values calculated after simulating lower coverage. This shows that ≥5× coverage is required. (d) The minimum required genome fraction for iRep was determined by conducting 100 random fragmentations and subsets of the L. gasseri genome. Sequencing was subset to 5× coverage before calculating iRep to show the combined affect of low coverage and missing genomic information. With ≥75% of a genome sequence, most iRep measurements are accurate ±0.15. (e) iRep and bPTR measurements were calculated using five genome sequences assembled from premature infant metagenomes, showing that these methods are in agreement in the context of microbiome sequencing data.