Abstract
A series of pyrazolo[3,4-d]pyrimidine derivatives related to allopurinol has been synthesized and evaluated for its cytotoxicity against a panel of three cancer cell lines as well as its xanthine oxidase (XOD) inhibitory activities. Among them, compound 4 showed potent cytotoxicity with IC50 values of 25.5 and 35.2 μM against human hepatoma carcinoma cell lines, BEL-7402 and SMMC-7221, respectively. The anticancer activity of 4 was comparable to that of Tanespimycin (17-N-allylamino-17-demethoxy geldanamycin, 17-AAG) that inhibited the growth of BEL-7402 and SMMC-7221 cells at IC50 values of 12.4 and 9.85 μM, respectively. However, unlike allopurinol, which is also a strong inhibitor of XOD, compound 4 is a much weaker XOD inhibitor, suggesting that the anticancer activities of the allopurinol derivatives may not be associated with XOD inhibition. Moreover, the cytotoxicity of 4 toward normal cells is significantly lower than that of 17-AAG, making 4 a promising lead compound for further optimization of structure-activity relationships that may lead to anticancer agents of clinical utility.
Keywords: allopurinol derivatives, cytotoxicity, human hepatoma carcinoma cell, XOD inhibition
1. Introduction
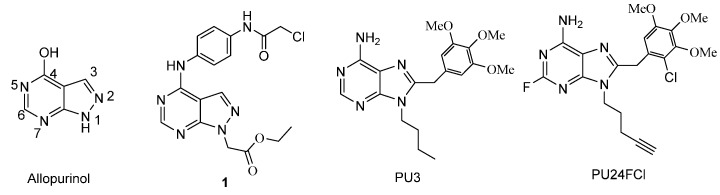
Allopurinol (4-hydroxyl-1H-pyrazolo[3,4-d]pyrimidine, Figure 1), a prototypical xanthine oxidase (XOD) inhibitor, has been used to treat hyperuricemia and its complications including chronic gout for several decades [1]. Yasuda et al. reported that allopurinol exerts cytotoxity on human hormone-refractory prostate cancer cells in combination with tumor necrosis factor-related apoptosis-inducing ligand [2]. Many other pyrazolo[3,4-d]pyrimidine derivatives [3] were extensively explored for their potential in the treatment of cancer by targeting casein kinase 1 (CK1) [4], hepatocyte growth factor receptor (c-MET) [5], erythropoietin-producing human hepatocellular receptors B (EphB) [6], vascular endothelial growth factor receptor 2 (VEGFR2) [6,7,8,9,10], Fms-like tyrosine kinase 3 (FLT3) [7,11], rearranged during transfection proto-oncogene (RET) [8,10], epidermal growth factor receptor (EGFR) [10], MARK (mitogen-activated protein kinases) signaling [9] and Wnt (Wingless-related integration site )/β-catenin signaling [11].
Figure 1.
Structures of allopurinol, compound 1, PU3, and PU24FCl.
Purine-based cancer therapeutics are well known in clinical applications [12]. Distinct from the traditional cytotoxic drugs, new types of purine analogs as anti-tumor drugs have been designed and developed to target the Hsp 90 protein involved in driving the cancer phenotype [13,14,15,16,17]. PU3 (Figure 1) is a lead compound for this type of small-molecule Hsp 90 inhibitor of purine analogs [13]. Optimization of PU3 led to PU24FCl, which exhibited wide-ranging anti-cancer activities at similar doses in all tested tumor types by specifically and potently inhibiting Hsp 90, while normal cells were 10- to 50-fold more resistant to these effects [14,18]. The successful design and development of PU3 and its simple analogs as anti-tumor agents suggest that further structural modifications to allopurinol might lead to novel analogs with improved pharmacological properties.
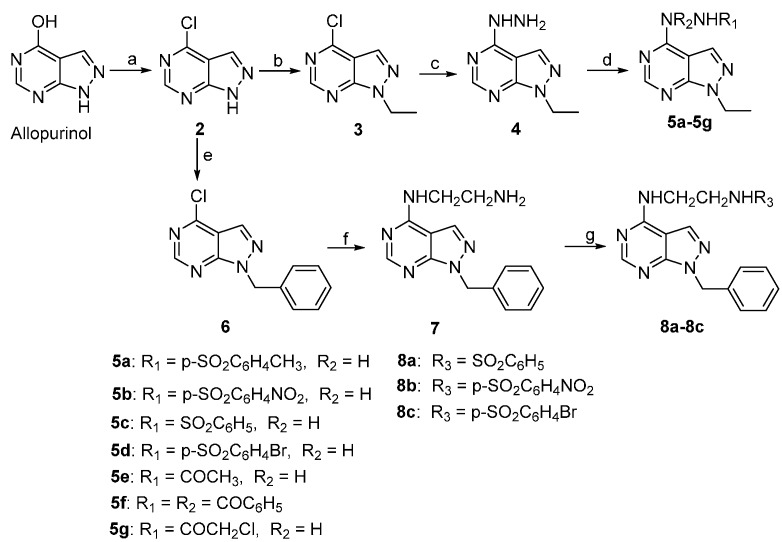
In a previous study, we chose allopurinol as a lead compound and synthesized a panel of pyrazolo[3,4-d]pyrimidine derivatives in the hope that some of the analogs may possess desirable anticancer properties [19]. However, only compound 1 of these derivatives showed potency against human hepatoma carcinoma cells 7402 and 7221 comparable to Tanespimycin (17-AAG), which is a heat shock protein 90 (Hsp 90) inhibitor and is currently in Phase II clinical trials [20]. It is also of note that, while the alkylating moiety present on the C-4 substituent of 1 may contribute to its potency, introduction of a carboxylic acid moiety as the N-1 substitution group appeared to abolish the activity entirely [19,21]. Thus, the dramatic loss of activity in all other allopurinol derivatives suggests that both the C-4 and N-1 substitutions are important in retaining cytotoxicity against cancer cells. In the present study, we are interested in exploring the effect of additional C-4 and N-1 substitution diversity on possible gains of anticancer activity in the resulting analogs. To this end, we have designed, synthesized, and evaluated a novel series of pyrazolo[3,4-d]pyrimidine derivatives where the C-4 moiety ranges from chlorine to unsubstituted and substituted hydrazines, and the N-1 substitution is either an ethyl or a benzyl group (Scheme 1).
Scheme 1.
Synthetic route of allopurinol derivatives. Reagents and conditions: (a) POCl3, (CH3)2NC6H5, 80 °C; (b) BrCH2CH3, triethylamine (TEA), KI, DMF, rt; (c) H2NNH2.H2O, CH3CN, rt; (d) RSO2Cl or RCOCl, pyridine, THF, 0 °C or TEA, CH2Cl2, 0 °C; (e) BrCH2C6H5, TEA, KI, DMF, rt; (f) H2NCH2CH2NH2, CH3CN, rt; and (g) RSO2Cl, TEA, DMF, 0 °C.
2. Results and Discussion
2.1. Chemistry
From commercially available allopurinol, compounds 2, 3, 4, 5a–5g, 6, 7 and 8a–8c were efficiently synthesized according to the synthetic route outlined in Scheme 1. Specifically, 4-chloro-1H-pyrazolo[3,4-d]pyrimidine (2) was obtained from the chlorination of allopurinol with POCl3. [22] N-Ethylation and benzylation of 2 with bromoethane or benzyl bromide under alkali conditions gave 4-chloro-1-ethyl-1H-pyrazolo[3,4-d]pyrimidine (3) and 1-benzyl-4-chloro-1H-pyrazolo[3,4-d]pyrimidine (6), respectively. The reaction of 3 with NH2NH2 affords 1-ethyl-4-hydrazinyl-1H-pyrazolo[3,4-d]pyrimidine (4). Next, acylation reactions of 4 with 4-methylbenzene-1-sulfonyl chloride, 4-nitrobenzene-1-sulfonyl chloride, benzenesulfonyl chloride, 4-bromobenzene-1-sulfonyl chloride or acetylchloride, benzoyl chloride, and chloroacetyl chloride lead to compounds 5a–5g, respectively. The mono-N-arylation of ethane-1, 2-diamine by 6 resulted in the formation of N1-(1-benzyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)ethane-1,2-diamine (7). Finally, compounds 8a–8c were formed by the acylation of 7 with benzenesulfonyl chloride, 4-nitrobenzene-1-sulfonyl chloride and 4-bromobenzene-1-sulfonyl chloride, respectively.
2.2. Biological Evaluation
2.2.1. Cytotoxicity against Cancer Cell Lines
We first tested the cytotoxic activities of the newly synthesized allopurinol derivatives in the two human hepatoma carcinoma cell lines, BEL-7402 and SMMC-7221. For comparison, 17-AAG was included as a positive control in the assay. As shown in Table 1, compound 4 emerges as the only derivative having comparable activity to 17-AAG and 1 among all allopurinol derivatives with IC50 values of 25.5 and 35.2 μM against the 7402 and 7221 hepatoma cell lines, respectively. In comparison, 17-AAG inhibited BEL-7402 and SMMC-7221 cells with IC50 of 12.4 μM and 9.85 μM, respectively. Importantly, the discovery of compound 4 as the only derivative possessing toxicity towards cancer cells revealed that alkylating moiety on C-4, while important, is not essential to an active allopurinol derivative. As observed with compound 4, when N-1 substitution is a simple ethyl group, a polar hydrazine on C-4 can bring about similar or greater cytotoxicity as an alkylating group in derivative 1. However, such cytotoxicity of allopurinol derivatives is rather sensitive to C-4 substitution, with a relatively narrow window of opportunity for desirable activity.
Table 1.
In vitro activities of allopurinol derivatives against human hepatoma carcinoma 7402 and 7221 cell lines.
| Compounds | IC50(μM) a | |
|---|---|---|
| BEL-7402 | SMMC-7221 | |
| 17-AAG | 12.4 ± 0.1 | 9.85 ± 0.08 |
| 1 | 18.7 [19] | 9.03 [19] |
| 3 | >100 b | >100 b |
| 4 | 25.5 ± 0.4 | 35.2 ± 0.4 |
| 5a | >100 b | >100 b |
| 5b | >100 b | >100 b |
| 5c | >100 b | >100 b |
| 5d | >100 b | >100 b |
| 5e | >100 b | >100 b |
| 5f | >100 b | >100 b |
| 5g | >100 b | >100 b |
| 6 | >100 b | >100 b |
| 7 | >100 b | >100 b |
| 8a | 66.61 ± 0.62 | >100 b |
| 8b | >100 b | 81.46 ± 0.78 |
| 8c | >100 b | >100 b |
17-AAG: Tanespimycin. a IC50 value represents the drug concentration required to inhibit cancer cell replication by 50%. b The compounds were tested up to a concentration of 100 μM.
When tested against another cancer cell line, MDA-MB-231, the allopurinol derivatives were found to exhibit moderate cytotoxicity. Shown in Table 2 are the relative survival rates of the analogs against MDA-MB-231 cells at 1 μM and 10 μM doses. For comparison, allopurinol, compound 1 and 17-AAG were included as references in the assay. Compound 4 reduced the survival rates of the cancer cells to 27.8% and 66.1% at 10 and 1 μM, again making it the most potent of all derivatives. While compound 4 is apparently more potent than allopurinol, it is less active than 1 and 17-AAG.
Table 2.
In vitro activity of allopurinol derivatives against MDA-MB-231 cell assays.
| Compounds | Survival Rate (%) | Compounds | Survival Rate (%) | ||
|---|---|---|---|---|---|
| 10 μM | 1 μM | 10 μM | 1 μM | ||
| 17-AAG | 4.8 ± 0.5 | 6.7 ± 0.5 | 5e | 61.3 ± 1.0 | 94.8 ± 0.5 |
| allopurinol | 51.7 ± 6.7 | 84.3 ± 7.7 | 5f | 88.1 ± 1.0 | 102.5 ± 11.0 |
| 1 | 1.0 ± 0.6 | 40.2 ± 3.9 | 5g | 105.4 ± 2.9 | 105.4 ± 4.8 |
| 3 | 67.0 ± 4.8 | 86.2 ± 8.6 | 6 | 75.7 ± 1.5 | 77.6 ± 9.1 |
| 4 | 27.8 ± 2.4 | 66.1 ± 0.5 | 7 | 57.5 ± 1.0 | 81.4 ± 7.2 |
| 5a | 72.8 ± 3.9 | 89.1 ± 1.5 | 8a | 58.4 ± 2.4 | 114..0 ± 9.1 |
| 5b | 68.0 ± 2.4 | 92.9 ± 1.5 | 8b | 71.8 ± 8.5 | 75.7 ± 2.4 |
| 5c | 74.7 ± 0.1 | 75.7 ± 11.0 | 8c | 78.5 ± 3.9 | 105.4 ± 8.6 |
| 5d | 70.9 ± 1.0 | 80.5 ± 2.0 | DMSO | 98.7 ± 6.3 | 100.6 ± 1.4 |
DMSO: Dimethylsulfoxide
2.2.2. Cytotoxicity towards MCF-10A Mammary Epithelial Cells
We next examined the cytotoxicity of the allopurinol derivatives in normal cells by treating the MCF-10A cells with compounds 4, 5e, and 5g at various concentrations. Again, compound 1 and 17-AAG were used as references for comparison. As shown in Table 3, compared to compound 1, whose activity is primarily attributable to its alkylating moiety, the new allopurinol derivatives exhibited significantly lower cytotoxicity than 1. At 17.36 µM of IC50, the new analog 4 is much less toxic in normal cells than that of 17-AAG at 0.09 µM IC50.
Table 3.
Cytotoxicity towards MCF-10A.
| Compounds | IC50 (μM) |
|---|---|
| 1 | 5.59 ± 0.27 |
| 4 | 17.36 ± 0.69 |
| 5e | 20.25 ± 0.81 |
| 5g | 23.29 ± 0.71 |
| 17-AAG | 0.09 ± 0.01 |
2.2.3. XOD Inhibitory Activity
XOD catalyzes the conversion of xanthine to uric acid. Since allopurinol is a classic XOD inhibitor, it is of interest to evaluate the allopurinol derivatives for their XOD inhibitory activity as a result of structural modifications to gain on anticancer activity. The XOD inhibition assay revealed that all tested allopurinol derivatives have lost the distinct inhibitory activity towards XOD, in sharp contrast to the potency exhibited by allopurinol as a positive control. Thus, the anticancer activity of the allopurinol derivatives does not appear to be associated with XOD inhibitory activity.
3. Materials and Methods
3.1. General Procedures for Chemical Synthesis
Allopurinol and xanthine oxidase were purchased from Sigma Aldrich (Shanghai, China). Dimethylsulfoxide (DMSO) was purchased from Amresco (Solon, OH, USA). All reagents were commercially available and were used without further purification unless otherwise indicated. Melting points were recorded with a micro melting point tester (Neware Technology Ltd., Guangdong, China) and are uncorrected. 1H and 13C-NMR spectra were obtained in DMSO-d6 or CDCl3 solutions on a Bruker AVII-400 spectrometer (Bruker, Fremont, CA, USA) operating at 400 and 100 MHz, respectively. The chemical shift values are given in δ units with reference to internal (CD3)4Si, and signals are designated as s (singlet), br s (broad singlet), d (doublet), t (triplet), q (quadruplet), dd (doublet of doublets), and m (multiplet), with coupling constants given in Hertz (Hz). High-resolution mass spectra were obtained on a Waters Q-TOF-Premiter instrument (TOF-MS) (Micromass, Manchester, UK). Infrared (IR) spectra were obtained by Perkin-Elmer 16PC-FT infrared spectrometer (Perkin-Elmer, Waltham, MA, USA). Thin layer chromatography (TLC) was employed to routinely monitor the reaction samples and confirm the homogeneity of the analytical samples by using Kieselgel 60F254 (0.25 mm) silica gel TLC aluminium sheets (Xiya Reagent, Chengdu, Sichuan, China). Column chromatography was carried out using silica gel 60, 200–300 mesh (Qingdao Haiyang Chemical, Qingdao, China).
3.1.1. 4-Chloro-1H-pyrazolo[3,4-d]pyrimidine (2)
A mixture of allopurinol (2.00 g, 14.69 mmol) and N,N-dimethylaniline (2.00 g, 16.52 mmol) was stirred in POCl3 (25 mL) at 80 °C for 2 h. The reaction mixture was diluted with water (35 mL), and extracted with ethyl acetate. The organic layer was washed with water and the organic phase was concentrated to dryness, and the residue was purified by column chromatography on silica gel using 4:1.5 petroleum ether/ethyl acetate as eluent to give 2 [22]: white flake solid, 70 % yield.
3.1.2. 4-Chloro-1-ethyl-1H-pyrazolo[3,4-d]pyrimidine (3)
To a solution of 4-chloro-1H-pyrazolo[3,4-d]pyrimidine 2 (0.1 g, 0.65 mmol) in anhydrous DMF (dimethylformamide, 5 mL ), TEA (trimethylamine, 0.20 g, 1.95 mmol) was firstly added, and mixture was stirred at room temperature for 30 min. Then, bromoethane (0.084 g, 0.78 mmol) was added and the mixture was stirred at room temperature for 1 h, and KI was then added. Then, the reaction mixture was diluted with water (15 mL), acidified with HCl and extracted with ethyl acetate. At last, the organic layer was washed with water and the organic phase was concentrated to dryness, and the residue was purified by column chromatography on silica gel using 10:1 petroleum ether/ethyl acetate as eluent to give 3: white powder, 76% yield, mp 76–77 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 8.87 (s, 1H, CH), 8.49 (s, 1H, CH), 4.51 (q, J = 7.24 Hz, 2H, CH2), 1.45(t, J = 7.26 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 156.7, 150.8, 150.4, 134.4, 106.1, 42.4, 15.1; IR (KBr, ν, cm−1): 3466, 3094, 2924, 2854, 1690, 1575, 1538, 1460, 1389, 1283, 1209, 1131, 1007, 956, 782, 680, 594, 533, 406. HRMS (ESI) calcd. for C7H8ClN4: [M + H]+ 183.0437, found 183.0535.
3.1.3. 1-Ethyl-4-hydrazinyl-1H-pyrazolo[3,4-d]pyrimidine (4)
To a solution of 4-chloro-1-ethyl-1H-pyrazolo[3,4-d]pyrimidine 3 (0.1 g, 0.56 mmol) in acetonitrile (5 mL), hydrazine hydrate(0.034 g, 0.68 mmol) was firstly added. Then, the mixture was stirred at room temperature for 30 min. Finally, the reaction mixture was concentrated to dryness, and the residue was purified by recrystallization using ethyl acetate to give 4: 75 % yield, mp 198–200 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 8.29 (s, 1H, CH), 8.05 (s, 1H, CH), 4.85 (s, 2H, NH2), 4.65 (s, 1H, NH), 4.30 (q, J = 7.18 Hz, 2H, CH2), 1.36 (t, J = 7.22 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 161.0, 155.4, 153.7, 135.2, 99.1, 41.5, 15.2; IR (KBr, ν, cm−1): 1717, 1659, 1599, 1538, 1497, 1441, 1375, 1346, 1295, 1251, 1182, 1091, 962, 913, 860, 785, 694, 620, 541. HRMS (ESI) calcd. for C7H11N6: [M + H]+ 179.1045, found 179.1044.
3.1.4. Synthesis of Compounds 5a–5g
To a solution of 1-ethyl-4-hydrazinyl-1H-pyrazolo[3,4-d]pyrimidine 4 (0.3 g, 1.69 mmol) in THF (5 mL), pyridine (0.19 g, 2.41 mmol) was firstly added at 0 °C and the mixture was stirred at 0 °C for 1 h. Then, substituted benzenesulfonyl chloride (2.05 mmol) or different acyl chloride was added and the reaction continued for 1 h at 0 °C. The reaction mixture was then diluted with water (15 mL), and extracted with ethyl acetate. At last, the organic layer was washed with water and the organic phase was dried over MgSO4 and concentrated, and the residue was purified by column chromatography on silica gel using 2:1 petroleum ether/acetone as eluent to give 5a–5g.
N′-(1-Ethyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)-4-methylbenzenesulfonohydrazide (5a): 61% yield, mp 186–187 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 10.28 (br, 1H, NH), 10.04 (br, 1H, NH), 8.16 (s, 2H, CH), 7.73 (d, J = 8.20 Hz, 2H, ArH), 7.41 (d, J = 8.08 Hz, 2H, ArH), 4.34 (q, J = 7.18 Hz, 2H, CH2), 2.39 (s, 3H, CH3), 1.37 (t, J = 7.20 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 161.2, 154.9, 153.2, 144.2, 135.8, 133.8, 130.0, 128.2, 99.5, 41.8, 21.5, 15.4; IR (KBr, ν, cm−1): 3437, 3194, 3052, 2924, 2855, 1923, 1592, 1499, 1454, 1336, 1292, 1245, 1162, 1091, 960, 917, 810, 789, 718, 658, 582, 538, 486, 423. HRMS (ESI) calcd. for C14H17N6O2S: [M + H]+ 333.1134, found 333.1128.
N′-(1-Ethyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)-4-nitrobenzenesulfonohydrazide (5b): 64% yield, mp 194–195 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 10.78 (br, 1H, NH), 10.17 (br, 1H, NH), 8.43 (s, 2H, CH), 8.17 (d, J = 8.76 Hz, 2H, ArH), 8.09 (d, J = 8.76 Hz, 2H, ArH), 4.34 (q, J = 7.24 Hz, 2H, CH2), 1.37 (t, J = 7.24 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 159.5, 157.9, 155.3, 152.6, 148.1, 138.5, 134.7, 129.6, 104.3, 46.6, 19.9; IR (KBr, ν, cm−1): 3364, 3262, 3106, 2625, 2855, 1698, 1657, 1597, 1530, 1449, 1350, 1312, 1242, 1171, 1088, 1010, 961, 855, 746, 703, 612, 462. HRMS (ESI) calcd. for C13H14N7O4S: [M + H]+ 364.0828, found 364.0837.
N′-(1-Ethyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)benzenesulfonohydrazide (5c): 65% yield, mp 194–195 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 10.40 (br, 1H, NH), 10.10 (br, 1H, NH), 8.16 (s, 2H, CH), 7.85 (d, J = 7.36 Hz, 2H, ArH), 7.60–7.69 (m, 3H, ArH), 4.33 (q, J = 7.12 Hz, 2H, CH2), 1.37 (t, J = 7.12 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 160.4, 154.9, 153.2, 138.8, 133.7, 129.6, 128.2, 99.6, 41.8, 15.2; IR (KBr, ν, cm−1): 3227, 3077, 2979, 2861, 1593, 1548, 1500, 1442, 1353, 1292, 1248, 1167, 1090, 1008, 959, 919, 857, 790, 739, 693, 637, 577, 542, 502. HRMS (ESI) calcd. for C14H15N6O2S: [M + H]+ 319.0977, found 319.0984.
4-Bromo-N′-(1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)benzenesulfonohydrazide (5d): 58% yield, mp 194–195 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 10.49 (br, 1H, NH), 10.09 (br, 1H, NH), 8.16 (s, 2H, CH), 7.76–7.84 (m, 4H, ArH), 4.34 (q, J = 7.18 Hz, 2H, CH2), 1.38 (t, J = 7.20 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 158.8, 154.9, 153.2, 137.7, 134.0, 132.7, 130.3, 127.8, 99.5, 41.8, 15.2; IR (KBr, ν, cm−1): 3361, 3259, 3185, 3130, 3090, 2924, 2854, 1913, 1658, 1598, 1565, 1537, 1388, 1318, 1282, 1159, 1087, 1066, 1007, 959, 907, 821, 759, 717, 674, 615, 575, 530. HRMS (ESI) calcd. for C13H14BrN6O2S: [M + H]+ 397.0082, found 397.0088.
N′-(1-Ethyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)acetohydrazide (5e): 54% yield, pale yellow oil, 1H-NMR (400 MHz, DMSO-d6) δ: 10.38 (br, 1H, NH), 10.09 (br, 1H, NH), 8.29 (s, 1H, CH), 8.16 (s, 1H, CH), 4.34 (q, J = 7.12 Hz, 2H, CH2), 2.03 (s, 3H, CH3), 1.37 (t, J = 7.20 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 170.1, 160.5, 155.4, 153.5, 132.3, 99.1, 41.8, 21.1, 15.2; IR (KBr, ν, cm−1): 3200, 3056, 2925, 2855, 1956, 1634, 1592, 1449, 1350, 1277, 1230, 1156, 1074, 1012, 891, 762, 708, 663, 597, 532. HRMS (ESI) calcd. for C9H13N6O: [M + H]+ 221.1151, found 221.1148.
N′-Benzoyl-N′-(1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)benzohydrazide (5f): 45% yield, mp 144–146 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 10.52 (br, 1H, NH), 8.67 (s, 1H, CH), 8.09 (s, 1H, CH), 7.92–7.96 (m, 4H, ArH), 7.80–7.82 (m, 2H, ArH), 7.74–7.76 (m, 2H, ArH), 7.68–7.70 (m, 2H, ArH), 4.49 (q, J = 7.30 Hz, 2H, CH2), 1.44 (t, J = 7.22 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 171.7, 171.0, 166.3, 154.6, 153.7, 134.6, 133.7, 133.0, 132.4, 129.0, 128.9, 128.0, 127.9, 104.6, 42.4, 15.1; IR (KBr, ν, cm−1): 3261, 3057, 2925, 2854, 1714, 1664, 1631, 1578, 1552, 1527, 1486, 1454, 1421, 1341, 1313, 1270, 1105, 958, 913, 869, 821, 795, 708, 691, 578, 545. HRMS (ESI) calcd. for C21H19N6O2 :[M + H]+ 387.1569, found 387.1577.
2-Chloro-N′-(1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)acetohydrazide (5g): 49% yield, mp 202–204 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 11.27 (br, 1H, NH), 10.28 (br, 1H, NH), 8.11–8.50 (m, 2H, CH), 4.37–4.41 (m, 4H, CH2), 1.41 (t, J = 7.20 Hz, 3H, CH3); 13C-NMR (100 MHz, DMSO-d6) 164.8, 150.4, 148.9, 146.3, 133.2, 96.4, 41.2, 40.1, 13.4; IR (KBr, ν, cm−1): 3428, 3134, 2949, 2629, 1722, 1663, 1611, 1499, 1403, 1345, 1223, 1106, 965, 884, 799, 700, 639, 527; HRMS (ESI) calcd. for C9H12ClN6O: [M + H]+ 255.0761, found 255.0794.
3.1.5. 1-Benzyl-4-chloro-1H-pyrazolo[3,4-d]pyrimidine (6)
To a solution of 4-chloro-1H-pyrazolo[3,4-d]pyrimidine 2 (0.1 g, 0.65 mmol) in anhydrous DMF (5 mL ), TEA (0.22 g, 0.65 mmol) was firstly added and stirred at room temperature for 30 min. Then, a solution of benzyl bromide (0.11 g, 0.78 mmol) in anhydrous DMF (5 mL) was added and the mixture was reacted for 1 h, then KI was added and continue reaction. The reaction mixture was at last diluted with water (15 mL), and extracted with ethyl acetate. The organic layer was washed with water and the organic phase was dried over MgSO4 and concentrated, and the residue was purified by column chromatography on silica gel using 10:1 petroleum ether/ethyl acetate as eluent to give 6: 65% yield, mp 62–63 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 8.91 (s, 1H, CH), 8.53 (s, 1H, CH), 7.26–7.33 (m, 5H, ArH), 5.70 (s, 2H, CH2); 13C-NMR (100 MHz, DMSO-d6) 156.6, 151.3, 150.9, 137.2, 135.1, 129.0, 129.0, 128.1, 128.1, 128.0, 106.3, 50.8; IR (KBr, ν, cm−1): 3441, 3090, 2935, 1897, 1756, 1585, 1547, 1479, 1410, 1347, 1243, 1174, 1135, 950, 858, 783, 730, 698, 596, 534; HRMS (ESI) calcd. for C12H10ClN4: [M + H]+ 245.0594, found 245.0584.
3.1.6. N1-(1-Benzyl-1H/-pyrazolo[3,4-d]pyrimidin-4-yl)ethane-1,2-diamine (7)
To a solution of 1-benzyl-4-chloro-1H-pyrazolo[3,4-d]pyrimidine 6 (0.10 g, 0.41 mmol) in acetonitrile (10 mL), ethane-1,2-diamine (0.025 g, 0.42 mmol) was firstly added and stirred at room temperature for 3 h. Then, the reaction mixture was concentrated to dryness, and the residue was purified by column chromatography on silica gel using 2:8 methylene chloride/methanol as eluent to give 7: 56% yield, yellow oil, 1H-NMR (400 MHz, DMSO-d6) δ: 8.61 (br, 1H, NH), 8.25 (s, 1H, CH), 8.03 (s, 1H, CH), 7.19–7.32 (m, 5H, ArH), 5.50 (s, 2H, CH2), 3.59 (m, 2H, CH2), 3.13 (m, 2H, CH2), 2.88 (t, J = 6.10 Hz, 2H, NH2); 13C-NMR (100 MHz, DMSO-d6) 156.5, 155.8, 152.7, 137.3, 132.4, 128.5, 127.4, 100.5, 49.7, 41.3, 40.6; IR (KBr, ν, cm−1): 3275, 2936, 2104, 1618, 1571, 1495, 1454, 1352, 1293, 1182, 1117, 1042, 918, 790, 732, 699; HRMS (ESI) calcd. for C14H17N6: [M + H]+ 269.1515, found 269.1506.
3.1.7. Synthesis of Compounds 8a–8c
To a solution of 7 (0.32 g, 1.19 mmol) in DMF10 mL), TEA (0.18 g, 0.78 mmol) was firstly added at 0 °C and stirred for 10 min. Then, substituted benzenesulfonyl chloride (1.34 mmol) was added and stirred at 0 °C for 2 h. The reaction mixture was diluted with water (15 mL), and extracted with ethyl acetate. At last, the organic layer was washed with water and the organic phase was dried over MgSO4 and concentrated. Then, the residue was purified by column chromatography on silica gel using 2:2:6 petroleum ether/ethyl acetate/methanol as eluent to give 8a–8c.
N-(2-((1-Benzyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)ethyl)benzenesulfonamide (8a): 61% yield, mp 114–115 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 8.35 (br d, J = 5.60 Hz, 1H, NH), 8.25 (s, 1H, CH), 8.08 (s, 1H, CH), 7.83 (t, J = 5.96 Hz, 1H, NH), 7.76–7.79 (m, 2H, ArH), 7.50–7.58 (m, 3H, ArH), 7.20–7.33 (m, 5H, ArH), 5.49 (s, 2H, CH2), 3.54 (m, 2H, CH2), 3.00 (m, 2H, CH2); 13C-NMR (100 MHz, DMSO-d6) 156.8, 156.2, 153.1, 140.7, 137.8, 132.8, 132.4, 129.6, 128.9, 128.0, 126.9, 101.0, 50.26, 42.2, 40.4; IR (KBr, ν, cm−1): 3539, 3400, 3325, 3148, 2925, 2862, 1614, 1572, 1537, 1492, 1450, 1380, 1305, 1153, 1087, 983, 903, 790, 727, 689, 647, 583; HRMS (ESI) calcd. for C20H21N6O2S: [M + H]+ 409.1447, found 409.1435.
N-(2-((1-Benzyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)ethyl)-4-nitrobenzenesulfonamide (8b): 66% yield, mp 171–172 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 8.30 (m, 3H), 8.21 (br, 1H, NH), 8.04 (s, 1H, CH), 7.99 (d, J = 8.72 Hz, 2H, ArH), 7.88 (br, 1H, NH), 7.23–7.31 (m, 3H, ArH), 7.19 (d, J = 7.24 Hz, 2H, ArH), 5.47 (s, 2H, CH2), 3.53 (m, 2H, CH2), 3.11 (m, 2H, CH2); 13C-NMR (100 MHz, DMSO-d6) 156.7, 156.2, 153.0, 149.7, 146.5, 137.7, 132.3, 128.9, 128.4, 127.9, 124.9, 100.9, 50.2, 42.1, 40.4; IR (KBr, ν, cm−1): 3405, 3232, 2925, 2854, 1611, 1570, 1528, 1494, 1437, 1378, 1352, 1345, 1313, 1274, 1252, 1169, 1067, 894, 855, 788, 739, 686, 651, 609, 559, 535, 462; HRMS (ESI) calcd. for C20H20N7O4S: [M + H]+ 454.1297, found 454.1292.
N-(2-((1-benzyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)ethyl)-4-bromobenzenesulfonamide (8c): 64% yield, mp 136–137 °C; 1H-NMR (400 MHz, DMSO-d6) δ: 8.35 (br, 1H, NH), 8.25 (s, 1H, CH), 8.09 (s, 1H, CH), 7.94 (br, 1H, NH), 7.72 (m, 4H, ArH), 7.25–7.32 (m, 3H, ArH), 7.21 (d, J = 6.96 Hz, 2H, ArH), 5.50 (s, 2H, CH2), 3.54 (m, 2H, CH2), 3.03 (m, 2H, CH2); 13C-NMR (100 MHz, DMSO-d6) 156.3, 155.71, 152.6, 139.6, 137.3, 132.2, 131.9, 128.5, 127.5, 126.1, 100.5, 49.8, 41.6, 40.1; IR (KBr, ν, cm−1): 3374, 2924, 2854, 1729, 1663, 1591, 1460, 1409, 1255, 1120, 1043, 854; HRMS (ESI) calcd. For C20H20BrN6O2S: [M + H]+ 487.0552, found 487.0562.
3.2. Biological Assays
3.2.1. Cell Culture
MDA-MB-231 cell line was routinely cultured in phenol red-free DMEM (Dulbecco’s modified eagle medium) medium supplemented with 5% FBS, 4 mM glutamine, 1 mM sodium pyruvate, 100 IU/mL penicillin, 100 μg/mL streptomycin and 0.25 μg/mL amphotericin. Cultures were maintained in 5% carbon dioxide at a temperature of 37 °C.
The Human Hepatoma Carcinoma Cell 7402 and 7221 cell lines were obtained from State Key of Biotherapy, Sichuan University, Sichuan, China. Logarithmically growing Human Hepatoma Carcinoma Cells 7402 and 7221 were incubated with 0.05% trypsin containing 1 mM EDTA at 37 °C for about 4 min until cells were non-adherent and formed a single cell suspension. Trypsin activity was neutralized by adding a 20-fold excess of serum-containing medium. Cells were cultured at 37 °C in an atmosphere of air and 5% CO2.
3.2.2. Cell Survival/Growth Assay
For growth assay in the presence of 10−5 M and 10−6 M individual compound, MDA-MB-231 cells were plated in six-well plates at a density of 50,000 per well in DMEM medium supplemented with 5% FBS. The cells were then cultured for 5 days, while equal treatment volumes of DMSO were used as the vehicle control. Cell numbers were counted with a Coulter instrument (Beckman-Coulter, Brea, CA, USA). The ratio of compound treated cell numbers to vehicle treated cell numbers was defined as survival ratio.
Cell viability of human hepatoma carcinoma cell 7402 and 7221 were assayed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). Briefly, human hepatoma carcinoma cell 7402 and 7221 were plated at a density of 1 × 105 cells/mL into 96-well plates and 7 × 103 cells/well plates, respectively. After overnight growth, cells were pretreated with a series of concentrations of acacetin for 24 h. The final concentrations of dimethyl sulfoxide in the culture medium were <0.1%. At the end of treatment, 10 μL of MTT was added, and cells were incubated for a further 4 h. Cell viability was determined by scanning with an ELISA reader with a 570 nm filter (BioTek, Winooski, VE, USA).
MCF-10A cells were purchased from ATCC (Manassas, VA, USA) and were maintained in DMEM/F12 medium containing 1.05 mM CaCl2, 100 mg/mL cholera toxin, 5% horse serum, 10 μg/mL insulin, 500 ng/mL hydrocortisone and 1% penicillin-streptomycin. The cells were cultured in cell culture incubators, which were set to temperatures of 37 °C and supplemented with 5% CO2. MCF-10A cells were plated at a density of 3 × 103 cells per well in 96-well plates, incubated overnight and then treated with serial dilutions of each individual drug using doses that closely corresponded to the individual IC50 values. After 72 h of exposure, the viable cell growth was measured using the Cell Titer-Glo luminescent cell viability assay kit (Promega, Madison, WI, USA).
3.2.3. Determination of Xanthine Oxidase (XOD) Inhibitory Activity
Measurement of XOD activity was carried out according to the reference method with slight modifications [23]. First, 1092 μL of 0.1 unit of XOD in buffer (200 mM sodium pyrophosphate/HCl, pH 7.5) and 2 μL (5, 10, 15, 25 μM) of the test extracts or compounds in DMSO were mixed at 37 °C for 5 min. The control group did not contain a test agent. The reaction was started by adding 200 μL of 0.6 mM xanthine in doubly distilled water to the mixture. The reaction mixture was incubated at ambient temperature. Finally, the absorption increments at 295 nm indicating the formation of uric acid were determined every minute up to 8 min. Allopurinol was used as a positive control. Three replicates were made for each test sample. The percent inhibition ratio (%) was calculated according to the following equation:
| % inhibition = [(rate of control reaction − rate of sample reaction)/rate of control reaction] × 100 | (1) |
4. Conclusions
A series of pyrazolo[3,4-d]pyrimidine derivatives from allopurinol were synthesized. In vitro cytotoxicity assays identified analog 4 as the most potent compound against breast cancer MDA-MB-231 and human hepatoma carcinoma BEL-7402 and SMMC-7221 cell lines. Its IC50 values of 25.5 and 35.2 μM are comparable to those of 17-AAG against BEL-7402 and SMMC-7221 cell lines, respectively. Importantly, the discovery of compound 4 with much greater potency than 5g sheds light on another venue of structural variation that can achieve cytotoxicity against cancer cells without the alkylating moiety. This finding thus provides strong support to the conclusion that the alkylating moiety in pyrazolo[3,4-d]pyrimidine derivatives of allopurinol, while important, is not essential to conferring anticancer activity. Compared with 17-AAG, the much lower cytotoxicity of 4 toward normal cells make it a promising lead compound for further development toward anticancer agents of desirable pharmaceutical profiles. Additional studies on anticancer mechanisms of 4 are under way in our laboratories.
Acknowledgments
The authors are thankful to the Analytical & Testing Center, Sichuan University, Sichuan, China for providing analytical data, and State Key Lab of Biotherapy, Sichuan University for completing the Human Hepatoma Carcinoma Cell 7402 and 7221 activity test. This project was supported by the Science and Technology Support Program of Sichuan Province (2014SZ0071) (Shu-Fan Yin) and in part by the NIH RCMI Cancer Research Center at Xavier University of Louisiana, City, State, Country through an NIMHD Grant 2G12MD007595 (Guangdi Wang).
Abbreviations
The following abbreviations are used in this manuscript:
- XOD
Xanthine oxidase
- 17-AAG
17-N-allylamino-17-demethoxy geldanamycin
- Hsp90
Heat Shock Protein 90
- TEA
Triethylamine
- DMF
Dimethylformamide
- DMEM
Dulbecco’s modified eagle medium
- FBS
Fetal bovine Serum
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NIH
National Institute of Health
- RCMI
Research Centers in Minority Institutions Program
- NIMHD
National Institute on Minority Health and Health Disparities
Author Contributions
Shu-Fan Yin conceived and designed the experiments; Yong Li, Ting-Ting Cao and Cai-Hu Li performed the synthetic experiments; Yong Li, Ying Li and Lin Dong analyzed the data; Shanchun Guo and Qiu Zhong evaluated the biological activity; Shilong Zheng and Guangdi Wang designed the biological experiments; Yong Li, Shu-Fan Yin, Shilong Zheng and Guangdi Wang wrote and revised the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Pacher P., Nivorozhkin A., Szabó C. Therapeutic effects of xanthine oxidase inhibitors: Renaissance half a century after the discovery of allopurinol. Pharmacol. Rev. 2006;58:87–114. doi: 10.1124/pr.58.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yasuda T., Yoshida T., Goda A.E., Horinaka M., Yano K., Shiraishi T., Wakada M., Mizutani Y., Miki T., Sakai T. Anti-gout agent allopurinol exerts cytotoxicity to human hormone-refractory prostate cancer cells in combination with tumor necrosis factor–related apoptosis-inducing ligand. Mol. Cancer Res. 2008;6:1852–1860. doi: 10.1158/1541-7786.MCR-08-0012. [DOI] [PubMed] [Google Scholar]
- 3.Martina Ferrari S., Motta C.L., Sartini S., Baldini E., Materazzi G., Politti U., Ruffilli I., Ulisse S., Miccoli P., Antonelli A. Pyrazolopyrimidine derivatives as antineoplastic agents: With a special focus on thyroid cancer. Mini Rev. Med. Chem. 2016;16:86–93. doi: 10.2174/1389557515666151016124208. [DOI] [PubMed] [Google Scholar]
- 4.Yang L.-L., Li G.-B., Yan H.-X., Sun Q.-Z., Ma S., Ji P., Wang Z.-R., Feng S., Zou J., Yang S.-Y. Discovery of N6-phenyl-1H-pyrazolo [3,4-d] pyrimidine-3,6-diamine derivatives as novel CK1 inhibitors using common-feature pharmacophore model based virtual screening and hit-to-lead optimization. Eur. J. Med. Chem. 2012;56:30–38. doi: 10.1016/j.ejmech.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Bannen L.C., Chan D.S.-M., Dalrymple L.E., Jammalamadaka V., Khoury R.G., Leahy G.W., Mac M.B., Mann G., Mann L.W., Nuss J.M., et al. C-met modulators and method of use. 7,977,345 B2. US Patent. 2011 Jul 12;
- 6.Holzer P., Imbach P., Furet P., Schmiedeberg N. Preparation of 3-(substituted amino)-pyrazolo[3,4-d]pyrimidines as Ephb4 receptors and VEGFR2 kinase inhibitors. 2007062805 A1. WO. 2007 Jun 7;
- 7.Yang L.-L., Li G.-B., Ma S., Zou C., Zhou S., Sun Q.-Z., Cheng C., Chen X., Wang L.-J., Feng S., et al. Structure-activity relationship studies of pyrazolo[3,4-d]pyrimidine derivatives leading to the discovery of a novel multikinase inhibitor that potently inhibits FLT3 and VEGFR2 and evaluation of its activity against acute myeloid leukemia in vitro and in vivo. J. Med. Chem. 2013;56:1641–1655. doi: 10.1021/jm301537p. [DOI] [PubMed] [Google Scholar]
- 8.Sartini S., Coviello V., Bruno A., La Pietra V., Marinelli L., Simorini F., Taliani S., Salerno S., Marini A.M., Fioravanti A. Structure-based optimization of tyrosine kinase inhibitor CLM3. Design, synthesis, functional evaluation, and molecular modeling studies. J. Med. Chem. 2014;57:1225–1235. doi: 10.1021/jm401358b. [DOI] [PubMed] [Google Scholar]
- 9.Chen X., Ji P., Yang H.-W., Yang L.-L., Zhou S., Zhong L., Ma S., Fu X.-Y., Zhou C., Li G.-B. Sc-535, a novel oral multikinase inhibitor, showed potent antitumor activity in human melanoma models. Cell. Physiol. Biochem. 2013;32:138–153. doi: 10.1159/000350123. [DOI] [PubMed] [Google Scholar]
- 10.Antonelli A., Bocci G., Motta C.L., Ferrari S.M., Fallahi P., Corrado A., Fioravanti A., Sartini S., Orlandi P., Piaggi S. CLM29, a multi-target pyrazolopyrimidine derivative, has anti-neoplastic activity in medullary thyroid cancer in vitro and in vivo. Mol. Cell. Endocrinol. 2014;393:56–64. doi: 10.1016/j.mce.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Ma S., Yang L.-L., Niu T., Cheng C., Zhong L., Zheng M.-W., Xiong Y., Li L.-L., Xiang R., Chen L.-J., et al. SKLB-677, an FLT3 and Wnt/β-catenin signaling inhibitor, displays potent activity in models of FLT3-driven AML. Sci. Rep. 2015;5 doi: 10.1038/srep15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pizzorno G., Diasio R.B., Cheng Y.C. Pyrimidine and purine antimetabolites. In: Kufe D.W., Pollock R.E., Weichselbaum R.R., Bast R.C.J., Gansler T.S., Holland J.F., Frei E.I., editors. Cancer Medicine. 6th ed. BC Decker; Hamiltom, ON, Canada: 2003. [Google Scholar]
- 13.Chiosis G., Timaul M.N., Lucas B., Munster P.N., Zheng F.F., Sepp-Lorenzino L., Rosen N. A small molecule designed to bind to the adenine nucleotide pocket of Hsp90 causes Her2 degradation and the growth arrest and differentiation of breast cancer cells. Chem. Biol. 2001;8:289–299. doi: 10.1016/S1074-5521(01)00015-1. [DOI] [PubMed] [Google Scholar]
- 14.Chiosis G., Lucas B., Shtil A., Huezo H., Rosen N. Development of a purine-scaffold novel class of Hsp90 binders that inhibit the proliferation of cancer cells and induce the degradation of Her2 tyrosine kinase. Bioorg. Med. Chem. 2002;10:3555–3564. doi: 10.1016/S0968-0896(02)00253-5. [DOI] [PubMed] [Google Scholar]
- 15.Chiosis G., Lucas B., Huezo H., Solit D., Basso A., Rosen N. Development of purine-scaffold small molecule inhibitors of Hsp90. Curr. Cancer Drug Targets. 2003;3:371–376. doi: 10.2174/1568009033481778. [DOI] [PubMed] [Google Scholar]
- 16.Wright L., Barril X., Dymock B., Sheridan L., Surgenor A., Beswick M., Drysdale M., Collier A., Massey A., Davies N. Structure-activity relationships in purine-based inhibitor binding to Hsp90 isoforms. Chem. Biol. 2004;11:775–785. doi: 10.1016/j.chembiol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 17.Dymock B., Barril X., Beswick M., Collier A., Davies N., Drysdale M., Fink A., Fromont C., Hubbard R.E., Massey A. Adenine derived inhibitors of the molecular chaperone HSP90-SAR explained through multiple X-ray structures. Bioorg. Med. Chem. Lett. 2004;14:325–328. doi: 10.1016/j.bmcl.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Vilenchik M., Solit D., Basso A., Huezo H., Lucas B., He H., Rosen N., Spampinato C., Modrich P., Chiosis G. Targeting wide-range oncogenic transformation via PU24FCl, a specific inhibitor of tumor Hsp90. Chem. Biol. 2004;11:787–797. doi: 10.1016/j.chembiol.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Yuan L., Song C., Li C., Li Y., Dong L., Yin S. Synthesis and biological evaluation of pyrazolo[4,3-d]pyrimidine analogues. Eur. J. Med. Chem. 2013;67:152–157. doi: 10.1016/j.ejmech.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Dimopoulos M.-A., Mitsiades C.S., Anderson K.C., Richardson P.G. Tanespimycin as antitumor therapy. Clin. Lymphoma Myeloma Leuk. 2011;11:17–22. doi: 10.3816/CLML.2011.n.002. [DOI] [PubMed] [Google Scholar]
- 21.Yang X., Sun G., Yang C., Wang B. Novel rhein analogues as potential anticancer agents. Chem. Med. Chem. 2011;6:2294–2301. doi: 10.1002/cmdc.201100384. [DOI] [PubMed] [Google Scholar]
- 22.Kubo K., Shimizu T., Ohyama S.-I., Murooka H., Iwai A., Nakamura K., Hasegawa K., Kobayashi Y., Takahashi N., Takahashi K. Novel potent orally active selective VEGFR-2 tyrosine kinase inhibitors: Synthesis, structure-activity relationships, and antitumor activities of N-phenyl-N′-{4-(4-quinolyloxy) phenyl} ureas. J. Med. Chem. 2005;48:1359–1366. doi: 10.1021/jm030427r. [DOI] [PubMed] [Google Scholar]
- 23.Cos P., Ying L., Calomme M., Hu J.P., Cimanga K., van Poel B., Pieters L., Vlietinck A.J., Berghe D.V. Structure-activity relationship and classification of flavonoids as inhibitors of xanthine oxidase and superoxide scavengers. J. Nat. Prod. 1998;61:71–76. doi: 10.1021/np970237h. [DOI] [PubMed] [Google Scholar]