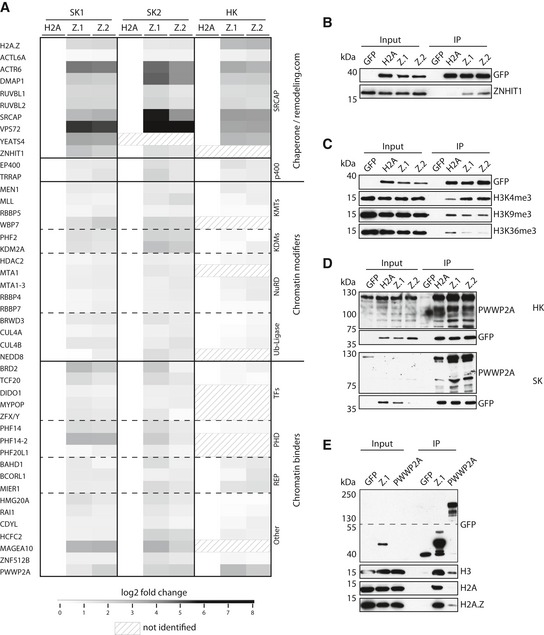
Figure 1. PWWP2A is a novel H2A.Z mononucleosome binder.
-
AHeatmap of significant outliers from pull‐downs analyzed by label‐free MS‐based proteomics with mononucleosomes derived from Skmel‐147 (two independent experiments: SK1 and SK2; see also Vardabasso et al, 2015 for first replicate) or HeLaK (HK) cells stably expressing GFP‐H2A (H2A), GFP‐H2A.Z.1 (Z.1), or GFP‐H2A.Z.2 (Z.2), normalized to H2A. Scale bar: log2‐fold. See also Fig EV1A–E for details on experimental procedure and verification of results, and Datasets EV1, EV2 and EV3 for detailed lists of H2A.Z‐binders.
-
BImmunoblotting of SRCAP complex‐specific member ZNHIT1 upon SK‐mel147 GFP, GFP‐H2A, GFP‐H2A.Z.1, or GFP‐H2A.Z.2 mono‐IPs. GFP served as control.
- C, D
-
EImmunoblots of GFP‐H2A.Z.1 or GFP‐PWWP2A mono‐IPs detecting endogenous H3, H2A, or H2A.Z. Notice enrichment of H2A.Z in comparison with H2A in GFP‐PWWP2A pull‐down. See also Fig EV1F–H for generation and characterization of GFP‐PWWP2A cell lines and endogenous pull‐down.
Source data are available online for this figure.
