Figure EV1. Negative stain EM structure of RERdj3WT reveals the native tetramer (related to Fig 2).
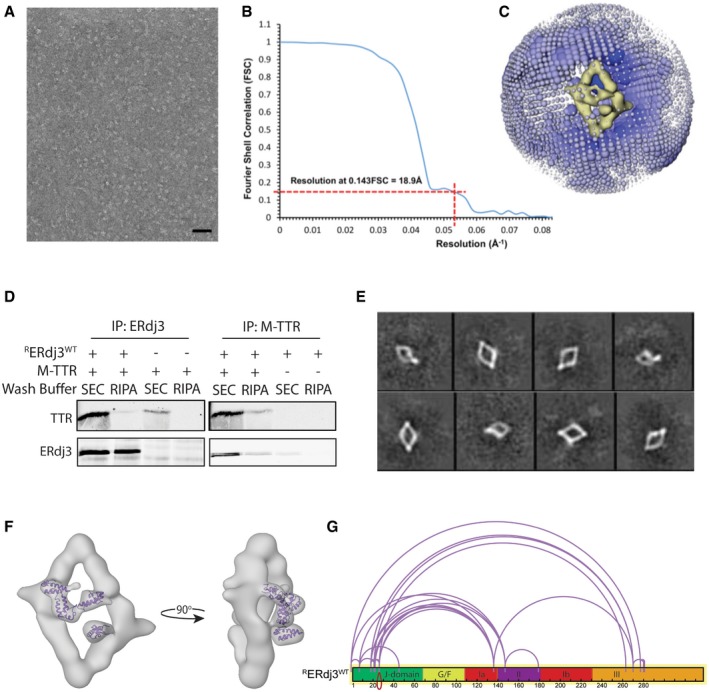
- A negative stain micrograph of recombinant ERdj3 (RERdj3; scale bar, 70 nm).
- Gold standard Fourier Shell Correlation (FSC) plot reporting the resolution of the final map to be 18.9 Å (at 0.143 FSC).
- Angular distribution of particles contributing to the 3D reconstruction, with the diameter of each sphere related to the number of particles at that Euler angle.
- Immunoblot of RERdj3 or monomeric transthyretin (M‐TTR) immunopurifications (IP) of solutions containing mixtures of RERdj3WT and M‐TTR, as indicated. The resulting IPs were washed with either the mild buffer used for size‐exclusion chromatography (SEC) or high‐detergent RIPA buffer, as indicated.
- 2D reference‐free class averages of negative stained RERdj3WT prepared in the presence of M‐TTR.
- EM image showing the NMR structure of DNAJB1 J‐domain (1HDJ; Qian et al, 1996) docked into the density localized above the ERdj3 tetramer reconstruction.
- Illustration showing the domain relationship for crosslinked peptides of RERdj3WT incubated in the presence of the amine‐reactive crosslinker DSS (10 μM). Crosslinked peptides were identified by liquid chromatography/mass spectrometry and mapped onto the RERdj3WT sequence using the xiNET crosslink viewer (Combe et al, 2015).
Source data are available online for this figure.
