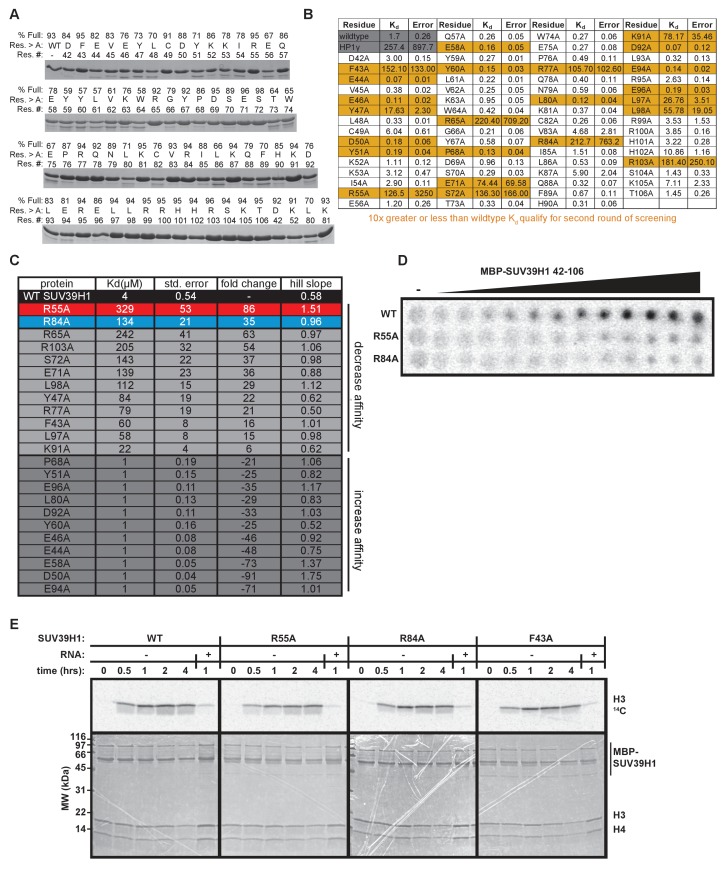
Figure 4. Identification and biochemical characterization of SUV39H1 RNA binding-deficient mutants.
(A) Identification of SUV39H1 mutants that affect RNA binding. Binding curves from second round of EMSA screening showing the binding of purified MBP-SUV39H1 42–106 point mutants to 19mer RNA (table of measured dissociation constants in Figure 4—figure supplement 1B). (B) Crystal structures of the chromodomains of human SUV39H1 (aa 44–91) (Wang et al., 2012) and human HP1α (aa 18–68) bound to H3K9me3 peptide (yellow) (Kaustov et al., 2011). Residues in SUV39H1 important for RNA binding (red and blue) or H3K9me3 binding (green and orange) are highlighted. The H3K9me3 peptide was co-crystalized with HP1α, but not SUV39H1. (C) Quantification of WT SUV39H1, SUV39H1R55A, and SUV39H1R84A binding to 19mer RNA. Binding curves generated by quantifying filter binding assays shown in Figure 4—figure supplement 1D. Error bars are standard deviation from three independent experiments. Dissociation constants (Kd, μM) displayed on graph are determined by non-linear fitting of the binding curves. Standard error represents the error of the curve fitting to the average of the three experimental replicates. (D) Coomassie stained gel of purified MBP-SUV39H1 42–106 proteins – WT, F43A, R55A, or R84A. (E) Binding of SUV39H1 mutants to H3K9me3. Representative α-MBP western blot showing the amount of purified MBP-SUV39H1 42–106 protein, WT or indicated mutant, bound to streptavidin beads conjugated to either H3K9me0 (me0), H3K9me3 (me3), or no peptide (-) added. (F) Quantification of western blot shown in 4E, error bars are standard deviation, n = 3, *p<0.03. See also Figure 4—figure supplement 1.