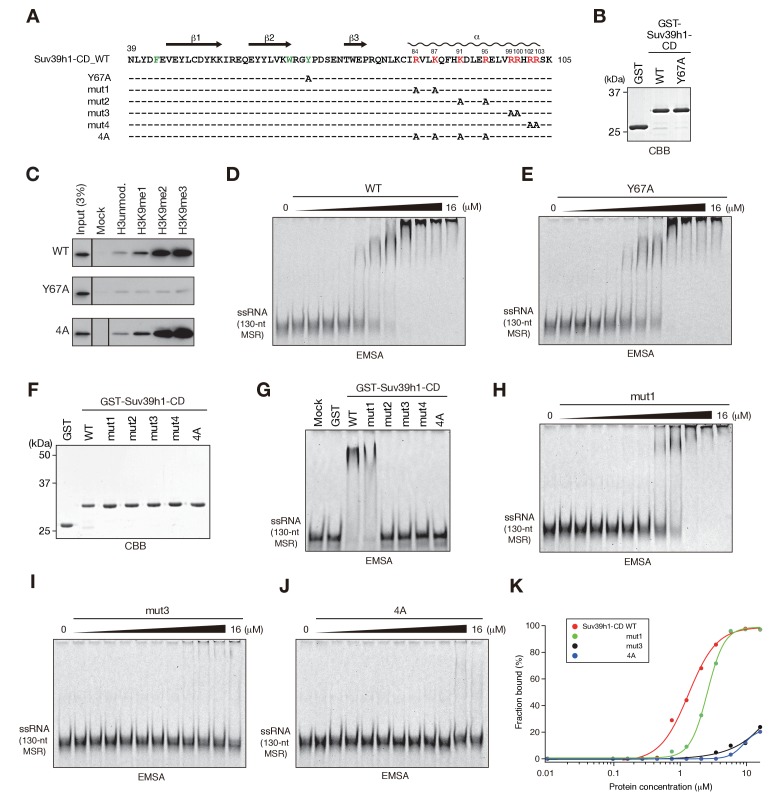
Figure 2. The residues required for Suv39h1-CD to bind RNA.
(A) The alignment of wild-type (WT) and mutant Suv39h1-CD (39–105). The upper lines indicate secondary structure. The positively charged amino acids in the C-terminal α-helix are indicated in red. The aromatic cage residues responsible for Suv39h1-CD’s recognition of H3K9me3 were indicated in green. (B, F) The recombinant proteins used in (C–E and G–J); the proteins were visualized by CBB staining. (C) An in vitro peptide-binding assay using wild-type (WT) and mutant (Y67A or 4A) Suv39h1-CDs. Biotin-tagged H3 (1–21) peptide (100 pmol) was incubated with GST-Suv39h1-CD (1 pmol), and the pulled-down proteins were analyzed by western blotting with an anti-GST antibody. (D, E, H–J) Titration EMSAs using serially diluted GST-fused WT or mutant Suv39h1-CD. (G) An EMSA using GST-fused WT or mutant Suv39h1-CD. (K) Binding isotherms of WT and mutant Suv39h1-CD proteins for ssRNA.