Abstract
The alveolar epithelium is composed of type I cells covering most of the gas-blood exchange surface and type II cells secreting surfactant that lowers surface tension of alveoli to prevent alveolar collapse. Here, we have identified a subgroup of type II cells expressing a higher level of cell surface molecule CD44 (CD44high type II cells) that composed ~3% of total type II cells in 5–10-wk-old mice. These cells were preferentially apposed to lung capillaries. They displayed a higher proliferation rate and augmented differentiation capacity into type I cells and the ability to form alveolar organoids compared with CD44low type II cells. Moreover, in aged mice, 18–24 mo old, the percentage of CD44high type II cells among all type II cells was increased, but these cells showed decreased progenitor properties. Thus CD44high type II cells likely represent a type II cell subpopulation important for constitutive regulation of alveolar homeostasis.
Keywords: CD44, alveoli, lung, type II cells, homeostasis
because the lung alveolar epithelium is in direct contact with the environment, it has a propensity for self-repair needed to maintain the integrity of alveoli in the basal state (37). The alveolar epithelium is composed of two types of cells, type I and type II cells (37). Both are derived from common progenitors located in the distal endoderm of the embryonic lung (4, 24). Through the onset of separate differentiation programs, these progenitors form these two cell types with distinct morphologies and functions (4, 24). Type I cells are flat and squamous, covering ~95% of the alveolar surface area, and provide the single-cell-thick interface between alveoli and underlining capillaries to enable efficient gas exchange (29). Type II cells are cuboidal and occupy only ~5% of alveolar surface area (23). Type II cells have multiple functions, including secretion of surfactant that prevents alveolar collapse (23) and modulation of lung immunity (23). In addition, studies using lineage-tracing analysis showed that type II cells self-renew and differentiate into type I cells following injury (6, 7).
In contrast to postinjury, the lung alveolar epithelial cell turnover rate is significantly slower in the steady state compared with the skin and the intestine (9, 11, 30, 36). Cell proliferation and type II-to-type I cell transition were observed only within small clusters of type II cells located at distinct foci in alveoli (1, 4). Alveolar type II cells have been classically regarded to be a homogeneous population (23). However, in recent years, subgroups of type II cells expressing distinct genes and with specialized functions have been identified (21, 26, 27, 33). For example, ~10% of type II cells express CC10, a gene highly expressed primarily in club cells in the airway (26). However, it is not known whether these cells harbor distinct progenitor properties. Heterogeneity of type II cells is clearly evident using single-cell sequencing techniques (33). Some type II cells upon injury showed a distinct genetic signature compared with other type II cells: hyperoxia exposure induced a subpopulation of type II cells expressing lower levels of E-cadherin with higher telomerase activities and enhanced proliferation compared with other type II cells (27). In Pseudomonas aeruginosa-induced pneumonia, a subpopulation of stem cell antigen-1-positive (Sca-1+) type II cells were identified having higher potential for differentiation into type I cells compared with Sca-1− counterparts (21, 22). Interestingly, these type II subgroups displayed a distinct progenitor property only during alveolar epithelial injury, raising the question of whether there is also a type II cell population responsible for maintaining alveolar epithelial homeostasis in the basal state. Here, we have identified such a population. These type II cells expressed a higher level of the transmembrane glycoprotein CD44 (41) and showed progenitor properties in uninjured lungs. CD44 is a receptor for hyaluronan (HA), a polysaccharide in the extracellular matrix (12), and it has been shown that CD44 has an altered expression pattern in fibrotic lungs (14). We found that in 5–10-wk-old mice, CD44high (or CD44hi) type II cells accounted for ~3% of total type II cells. Compared with their CD44low (or CD44lo) counterparts, CD44high type II cells showed a higher proliferation rate and greater tendency to differentiate into type I cells and to form alveolar organoids in three-dimensional (3-D) culture. Moreover, in aged mice, there was an increase in the percentage of CD44high type II cells, but these cells displayed decreased progenitor cell properties compared with those from younger mice. These results point to a role of CD44high type II cells in replacing lost alveolar epithelial cells and thereby contributing to steady-state alveolar homeostasis.
MATERIALS AND METHODS
Mice.
All animal studies were approved by the Institutional Animal Care Committee of the University of Illinois at Chicago. SpC-CreER/Rosa-Tomato and SpC-CreER/Rosa-mTmG mice were generated by crossing the Sftpc-CreERT2 mouse line (28; abbreviated SPC-CreER and kindly provided by Dr. Brigid Hogan) with the B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (The Jackson Laboratory) or B6.129(Cg)-Gt(ROSA)26Sortm4(ACTBtdTomato,-EGFP)Luo/J lines (The Jackson Laboratory), respectively. We chose either of these two lines on the basis of convenience when combining with other fluorescent labels. SpC-CreER/Rosa-tTA/tetO-H2B-GFP were generated by crossing the Sftpc-CreERT2 mouse line with the STOCK Tg(tetO-HIST1H2BJ/GFP)47Efu/J (The Jackson Laboratory) and then with the B6.129P2(Cg)-Gt(ROSA)26Sortm1(tTA)Roos/J (The Jackson Laboratory) line. Tamoxifen (Sigma-Aldrich, St. Louis, MO) was administered four times at days 0, 2, 4, and 6 by intraperitoneal injection at a dose of 0.25 mg per gram of mouse weight for each injection (21, 28). Tamoxifen-injected mice were then maintained for 2–4 wk before use in experiments. CD44null mice [B6.129(Cg)-CD44tm1Hbg/J; 25] and C57BL/6 mice were from The Jackson Laboratory. Bromodeoxyuridine (BrdU; Sigma-Aldrich) was injected intraperitoneally into CD44null and C57BL/6 mice at 75 mg/kg body wt each day for 1 wk. All mice were 5–10 wk old unless otherwise indicated.
Immunohistochemistry.
For histological analysis, lungs were perfused with 10–20 ml of PBS through the right ventricle before fixing in 4% paraformaldehyde (PFA; injected through the trachea) overnight at 4°C (21). Paraffin sections of fixed lungs (5 µm thick) were prepared at the Histology Core at the University of Illinois at Chicago. The following antibodies were used in this study: rat anti-mouse CD44 (1:50; BioLegend), rabbit anti-surfactant protein-C (anti-Sp-C; 1:500; Millipore), chicken anti-green fluorescent protein (anti-GFP; 1:500; Aves Laboratories), hamster anti-T1α (1:50; Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa), goat anti-Sp-C (1:50; Santa Cruz Biotechnology), rabbit anti-HOP homeobox (anti-HopX; 1:50; Santa Cruz Biotechnology), rabbit anti-von Willebrand factor (anti-vWF; 1:100; Chemicon), and rat anti-receptor for advanced glycation end products (anti-RAGE; 8–25 µg/ml; R&D Systems). Fluorescent secondary antibodies were from Jackson Immunoresearch and diluted 1:200. Images were taken using a Zeiss confocal microscope (LSM-880; Carl Zeiss, Oberkochen, Germany). Controls and experimental images were always taken with the same exposure parameters. If adjustment of images (brightness or contrast) was involved, the degrees of adjustment were always the same for control and experimental images.
Isolation of alveolar epithelial type II cells and flow cytometry.
Type II cells were isolated as previously described (5, 22). Briefly, PBS-perfused lungs were digested with Dispase (injected through the trachea; Corning) at room temperature for 45 min. The cell suspension was then treated with DNase I and sequentially filtered through a 70-μm cell strainer and 20-μm nylon gauze (Small Parts). Endothelial and immune cell contamination was minimized by panning cells on plates coated with anti-CD45 and anti-CD32 antibodies (BioLegend). Type II cells were pelleted by centrifugation for 6 min at 150 g. We further increased the purity of type II cells by fluorescence-activated cell sorting (FACS) using the above SpC-CreER-driven lineage-tracing mice or using epithelial cell adhesion molecule (EpCAM) as a selection marker to ensure that the cells we analyzed were over 95% purity for type II cells (21).
For flow cytometry, freshly isolated type II cells were blocked with 3% BSA in PBS. Antibodies were used as follows: allophycocyanin (APC)-CD44 (BioLegend), FITC-EpCAM (BioLegend), FITC- or phycoerythrin (PE)-cyanine 7 (Cy7)-Sca-1 (BioLegend), and FITC-integrin-β4 and FITC-integrin-α6 (BioLegend). After antibody staining, cells were briefly counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to eliminate dead cells from the analysis. Cells were sorted using a Beckman Coulter MoFlo high-speed cell sorter or analyzed with a CyAn ADP flow cytometer (Beckman Coulter), both located at the Flow Cytometry Core of Research Resources Center at the University of Illinois at Chicago. For each FACS experiment, isotype and single color controls were performed, and the gates for measuring CD44 or other markers were set on the basis of the results of isotype controls and single color controls conducted at the same time. The results were analyzed using Cytomation Summit 6.2 software (Beckman Coulter).
In vitro culture.
For most 2-D cultures, 10,000 FACS-sorted type II cells were cultured in gelatin-coated 48-well plates or chamber slides. For some experiments, plates or chamber slides were coated with collagen at 6 μg/cm2 overnight. Cells were cultured in 200 µl of DMEM supplemented with 10% FBS, penicillin/streptomycin, and 25 mM HEPES. The plates or slides were further processed for immunostaining.
For 3-D culture, 5,000 sorted type II cells and 1 × 105 Mlg mouse fibroblast cells (American Type Culture Collection, Manassas, VA) were resuspended in 200 µl of serum-free medium containing 80 µl of growth factor-reduced Matrigel (Corning) and 20 µl of rat tail collagen (Fisher Scientific). The Matrigel/cell mixture was seeded in a 24-well insert (Corning) placed in a 24-well plate with 600 μl of medium added to the outside of the insert. DMEM/F-12 (Invitrogen, Carlsbad, CA) supplemented with 10% FBS, penicillin/streptomycin, 1 mM HEPES, and insulin/transferrin/selenium (Sigma Chemical) was used in 3-D culture. The medium was changed after the first day of culture and then every other day for 2 wk (1, 20). Fluorescent images of colonies formed in the 3-D culture were taken using an EVOS FL microscope (Thermo Fisher Scientific). Later, the Matrigel plugs were fixed with 4% PFA/PBS overnight and separated from the inserts using forceps, before being paraffin embedded and sectioned in the Histology Core at the University of Illinois at Chicago. For some experiments, high-molecular weight (HMW) HA (Abbott) was added at 200 µg/ml in Matrigel mixture and medium. The sizes of the colonies were measured with ImageJ software.
Real-time PCR.
RNA from FACS-sorted cells was extracted using the RNAqueous-Micro Total RNA Isolation Kit (Ambion) and reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The resulting cDNA was amplified using the FastStart Universal SYBR Green Master Kit (Roche Life Science) and analyzed on the ABI ViiA7 system (Applied Biosystems). Primers for CDC25C and CCNB1 were obtained from Qiagen. CD44 total, CD44s, and CD44 v8/v9 primers were kindly provided by Dr. Chonghui Cheng (2). Gene expression was normalized to the level of cyclophilin as an internal control (22, 40).
Single-cell RNA-sequencing data analysis.
We searched GSE52583 (for mouse cells) and GSE86618 (for human cells) from Gene Expression Omnibus database from National Center for Biotechnology Information and downloaded GSE52583_RAW.tar and GSE86618_34816genes-540cells-tpm.txt.gz, respectively. For the mouse study, the data set consists of processed expression data [expressed as fragments per kilobase of transcript per million mapped reads (FPKM)] from single-cell RNA sequencing (RNAseq) of 198 mouse lung epithelial cells at 4 different developmental stages. From these we pulled out the results of the 46 adult type II cells and analyzed CD44 expression levels in these cells. CD44 levels (FPKM) of each of the 46 type II cells were copied into an Excel form and plotted. For human studies, the data set consists of processed expression data [presented as transcripts per kilobase million (TPM)], of single-cell RNAseq of type II cells isolated from idiopathic pulmonary fibrosis patient samples and normal transplant donor samples. We analyzed only the normal human samples (from donors cc019, cc002, and cc006). CD44 levels as well as the levels of Sp-C, Sp-B, and Sp-A (in TPM) of each cell from donors cc019, cc002, and cc006 were recorded on an Excel form, the non-type-II cells were excluded on the basis of low Sp-C level, and CD44 levels of each of the type II cells were plotted.
Statistical analysis.
Values of different groups were calculated using Microsoft Excel and were compared by Student’s t-test for statistical significance. P < 0.05 was regarded as statistically significant. Box-whisker plots were drawn (using GraphPad Prism 5.01 software) as standard Tukey box plots. In a box plot, the upper and lower boundaries of the box correspond to the first and third quartiles, and the line in the middle of the box is plotted at the median. The upper and lower whiskers are the highest and lowest values that are within 1.5× interquartile range (IQR) from the box. Values beyond the 1.5× IQR range are plotted as individual dots.
RESULTS
Lineage-tracing identification of CD44high type II cells in the adult mouse lung.
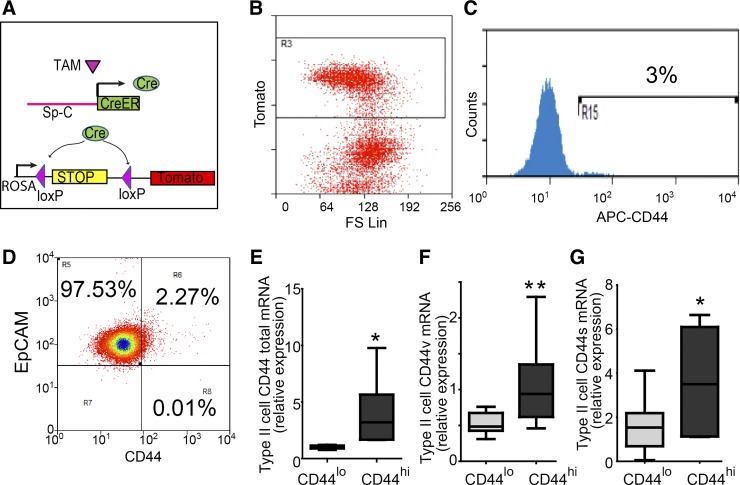
To identify subpopulations of alveolar type II cells showing progenitor cell properties and contributing to the maintenance of alveolar epithelium, we examined the presence of cell surface markers on type II cells isolated from adult wild-type mice. Type II cells specifically express surfactant protein-C (Sp-C; 1, 4), which can be labeled by SpC-CreER/Rosa-Tomato or SpC-CreER/Rosa-mTmG reporter mice (1, 4). In these lines, Cre recombinase is expressed only through the type II cell-specific Sp-C promoter after induction by tamoxifen. Cre cleaves DNA fragments flanked by loxP sites to enable the expression of the fluorescent lineage-tracing markers Tomato (for SpC-CreER/Rosa-Tomato line; Fig. 1A) or GFP (for SpC-CreER/Rosa-mTmG line; 1, 4) in Sp-C+ type II cells and their progenies. Alveolar type II cells were isolated from tamoxifen-treated lineage-tracing mice (5–10 wk old) and subjected to fluorescence-activated cell sorting (FACS) analysis. We found that 70% of the isolated type II cells were lineage labeled (Fig. 1B), consistent with previous studies (1, 21). From FACS analysis of lineage-labeled type II cells, we found that 1–4% of these cells expressed the stem cell marker CD44 (41; Fig. 1, B and C). In addition, almost all (>99.6%, Fig. 1D) of the lineage-labeled CD44high cells were positive for epithelial cell marker EpCAM (21), indicating that the lineage-labeled line is highly specific.
Fig. 1.
Isolation of CD44high type II cells from Sp-C lineage-tracing mouse lines. A: to identify type II cells from adult mice, four doses of tamoxifen (TAM; 0.25 mg/g) were given to SpC-CreER/Rosa-Tomato mice, in which TAM-induced Cre activation causes excision of the stop codon upstream of Tomato, resulting in lineage labeling of Sp-C+ cells. B and C: alveolar epithelial cells were isolated using a type II cell-enriched protocol and subjected to FACS analysis. Type II cells were selected by gating for the Tomato+ population (B) and were further separated into CD44low and CD44high populations (C). Approximately 3% of the type II cells were positive for CD44. FS Lin, forward scatter linear; R3, region 3. D: Tomato+ cells were gated and analyzed for the expression of EpCAM and CD44. Of the Tomato+ cells, 99.8% were EpCAM+, and of the Tomato+ CD44high cells, 99.6% were EpCAM+. These data are representative of >5 independent experiments. E–G: expression levels of CD44 isoforms in sorted cells were analyzed using qRT-PCR using primers that specifically detect all isoform types (CD44 total; E), the variant isoforms containing variable exons (CD44v; F), or standard isoform (CD44s; G). Compared with CD44low type II cells, CD44high type II cells expressed significantly higher levels of total CD44 (E), CD44v (F), and CD44s (G). Data are presented as Tukey box plots. *P < 0.05, **P < 0.01; n = 7 mice.
We further compared the transcription level of CD44 between the CD44high and CD44low type II cells. Tomato+CD44high and Tomato+CD44low cells were isolated from SpC-CreER/Rosa-Tomato mice using FACS and subjected to quantitative reverse transcription PCR (qRT-PCR) analysis. The expression level of total CD44 was significantly higher in CD44high cells (Fig. 1E). Because CD44 consists of variable exons, cells can produce CD44 isoforms through alternative splicing (2), we further analyzed the expression levels of variable exon-containing isoforms (CD44v) and the standard isoform (CD44s), in the FACS-isolated cells (2). Using CD44v primers, which amplify isoforms containing variable exons v8 and v9, and CD44s primers, which amplify the standard isoform containing no variable exon (2), we found that both CD44v and CD44s were expressed higher in CD44high type II cells compared with their CD44low counterparts (Fig. 1, F and G).
CD44high type II cells show higher proliferation rates.
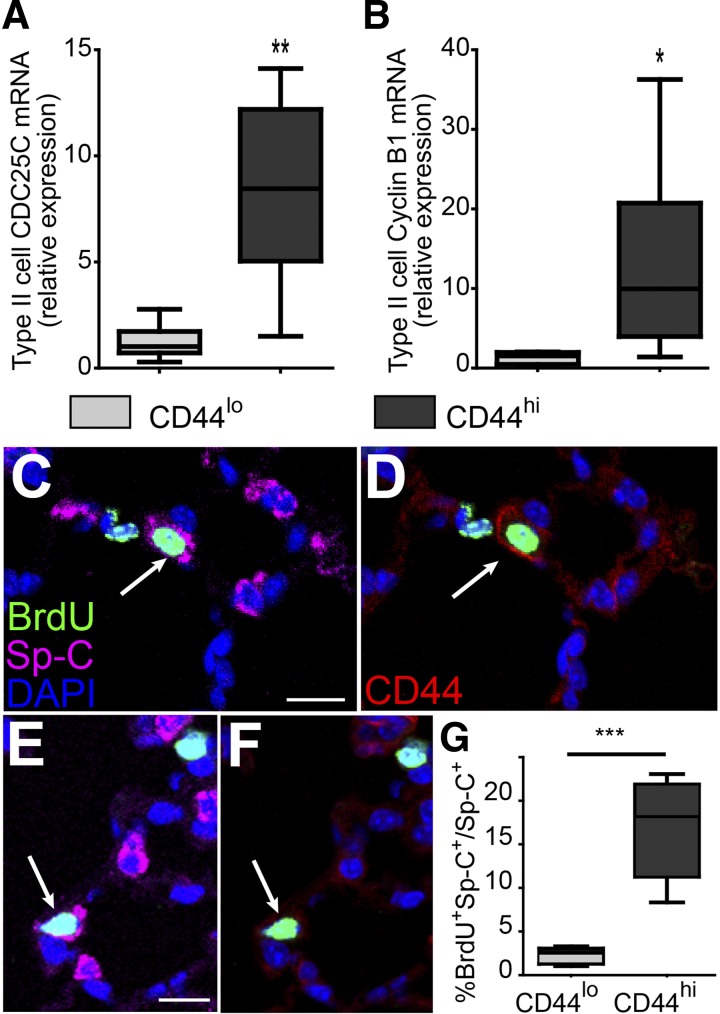
Because CD44 is a surface marker of various stem cells (10, 31, 41), we next examined whether the identified CD44high type II cells showed progenitor cell characteristics, such as self-renewal and differentiation into type I cells. The proliferation potential of freshly isolated Tomato+CD44high and Tomato+CD44low cells was determined by comparing the expression levels of CDC25C and cyclin B1 (CCNB1), cell cycle genes expressed in proliferating type II cells (21). Both genes showed significantly higher levels of expression in CD44high cells compared with CD44low cells (Fig. 2, A and B), indicating a higher proliferation rate of CD44high type II cells compared with CD44low type II cells.
Fig. 2.
CD44high type II cells show higher cell proliferation compared with CD44low type II cells. A and B: relative expression of CDC25C (A) and cyclin B1 (CCNB1; B) in sorted CD44high and CD44low type II cells was analyzed using qRT-PCR. Both genes showed significantly higher expression levels in CD44high type II cells compared with CD44low type II cells. Data are presented as Tukey box plots. *P < 0.05, **P < 0.01; n = 7 mice. C–G: BrdU-labeled type II cells in C57BL/6 mice. Lungs from BrdU-injected mice were subjected to immunohistological analysis, and cell proliferation rates of CD44low or CD44high type II cells were scored as BrdU+Sp-C+CD44low cells vs. Sp-C+CD44low cells or BrdU+Sp-C+CD44high cells vs. Sp-C+CD44high cells, respectively. C and D show the same section with a BrdU+Sp-C+CD44high cell, and E and F show the same section with a BrdU+Sp-C+CD44low cell. G: for each mouse, ~600 Sp-C+ cells were scored; n = 5 mice. Data are presented as Tukey box plot. ***P < 0.001.
To validate the qRT-PCR results, BrdU was injected into 5–10-wk-old C57BL/6 mice to label proliferating type II cells in the lung. Because of slow turnover of alveolar epithelial cells during steady-state conditions (1, 4), BrdU was injected for 1 wk at 75 mg/kg body wt each day. Lungs were then subjected to immunohistological analysis, and the ratios of BrdU+Sp-C+CD44low cells vs. Sp-C+CD44low cells as well as BrdU+Sp-C+CD44high cells vs. Sp-C+CD44high cells were scored by counting the cells from randomly selected areas on lung sections. We found that the fractions of BrdU-labeled cells were significantly greater among Sp-C+CD44high cells compared with Sp-C+CD44low cells (Fig. 2, C–G).
CD44high type II cells show enhanced differentiation in vitro.
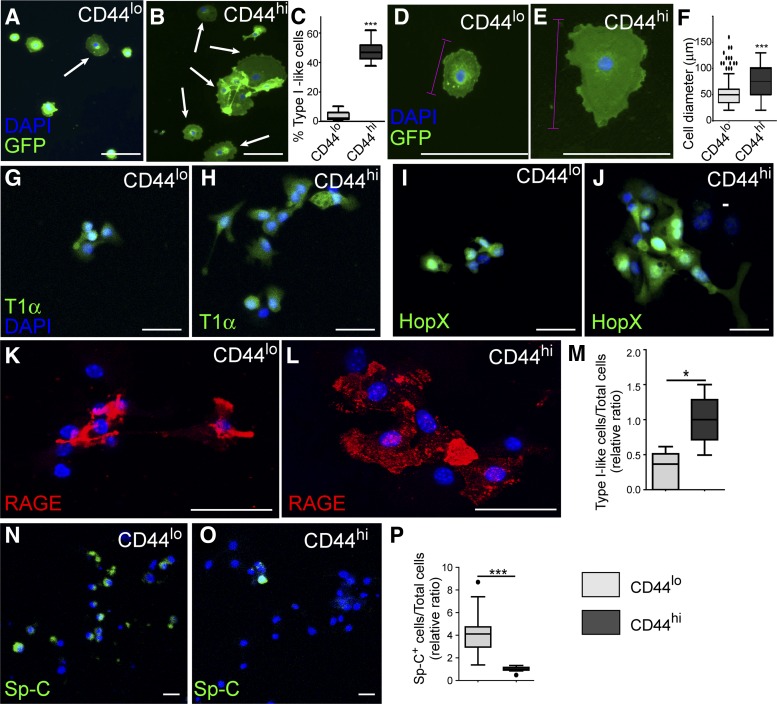
Next, we cultured CD44high and CD44low type II cells on gelatin-coated plates. At day 2 after culture initiation, some of the CD44high and CD44low type II cells started to change their shape to flattened squamous type I cell-like morphology. We found that the percentage of flat type I-like cells in the CD44high cell culture was greater than that of CD44low culture (Fig. 3, A–C). We also found that type I-like cells generated by CD44high type II cells were more elongated and appeared to spread into a larger area than CD44low cells (Fig. 3, D–F). Furthermore, when CD44high or CD44low cells were cultured on collagen-coated plates, they showed morphological transition behaviors similar to those of cells on gelatin-coated plates (Chen Q and Liu Y, unpublished observations).
Fig. 3.
CD44high type II cells differentiate into type I cells in vitro. CD44high and CD44low type II cells were grown on 0.2% gelatin-coated plates. A and B: after 48 h of culture, some cells adopted flattened type I cell-like morphology (arrows). C: percentages of the flattened type I-like cells were significantly higher in the CD44high cell cultures compared with those of CD4low cells. D and E: higher-magnification images of representative cells derived from CD44low (D) or CD44high (E) type II cells after growing 5 days in vitro. Cell diameters were measured on the basis of the longer axis of the cell (pink lines). F: quantification of cell diameters after 5 days of culture showed that average diameter of the flattened cells derived from CD44high type II cells were significantly larger than those from CD44low type II cells. G–L: cells were fixed at 72-h postculture and subjected to immunohistochemistry analysis of type I cell markers T1α (G and H), HopX (I and J), and RAGE (K and L). Some of the flattened-shaped cells from both CD44low (G, I, K) and CD44high (H, J, and L) cultures expressed these type I cell markers. M: cells expressing any of the above type I cell markers were counted, and the ratio of these type I-like cells to total cells was calculated. N and O: cells were stained for type II cell marker Sp-C at 72-h postculture. CD44low cells (N) had more Sp-C-expressing cells than CD44high cells did (O). P: Sp-C+ cells were counted, and the ratio of Sp-C+ cells to total cells was calculated. Scale bar = 30 µm for A, B, G–L, N, and O. Scale bar = 100 µm for D and E. The data in C, F, M, and P are presented as Tukey box plots. *P < 0.05, ***P < 0.001. Each sample group of the quantification data (C, F, M, and P) was from >5 mice; 50–200 cells randomly selected from each group from each mouse were scored. Because of the slight variation in culture condition of each independent experiment, M and P are plotted as the relative value compared with the average result of CD44high cells cultured at the same time.
Next, we stained the 3-day-cultured cells with type I cell markers T1α, HopX, and RAGE (39; Fig. 3, G–L). We found that some of the type I-like cells from both CD44high and CD44low culture expressed these markers. Next, the percentages of type I-like cells were further quantified on the basis of marker expression. Consistent with the morphology (Fig. 3C), there was a higher fraction of cells expressing type I cell markers in the CD44high cell culture compared with that of CD44low culture (Fig. 3M). In contrast, more of the cells in CD44low culture maintained the expression of type II cell marker Sp-C compared with that of CD44high cells (Fig. 3, N–P). Thus CD44high type II cells had a higher potential of converting into type I cells in vitro.
CD44high type II cells better form alveolar organoids in 3-D culture than CD44low cells.
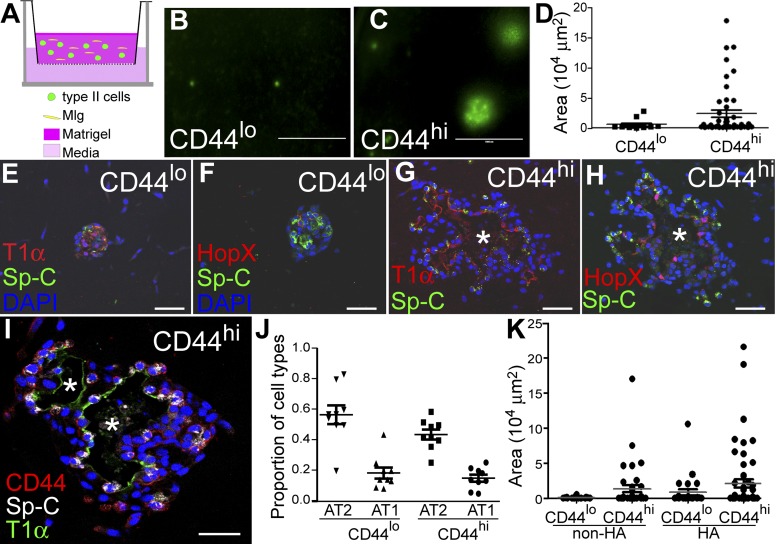
Next, we utilized a 3-D air-liquid interface organoid culture to better mimic the physiological environment of alveoli (1, 20). In this culture, the alveolar epithelial cells were cocultured with mesenchymal cells that serve as the progenitor cell niche (1, 20; Fig. 4A). Here, we used the lineage-tracing line SpC-CreER/Rosa-mTmG, in which type II cells and their progenies were labeled with GFP. We isolated GFP+CD44high and GFP+CD44low type II cells using FACS. Five thousand cells of each type were mixed with 1 × 105 neonatal mouse lung fibroblast Mlg cells (20) and cultured in a Matrigel/collagen mixture in Transwell inserts (Fig. 4A). After 2 wk of culture, a greater number of GFP+ colonies were formed with CD44high cells compared with CD44low cells. Colony forming efficiencies for CD44high and CD44low type II cells were 0.48 ± 0.03 and 0.12 ± 0.11% (means ± SD), respectively. The size of colonies also varied between CD44high and CD44low cell cultures; most colonies formed by CD44low cells were small (section area <10,000 μm2), and a few were medium sized (section area between 10,000 and 50,000 μm2). In contrast, large colonies (section area >50,000 μm2) were frequently detected in CD44high cultures (Fig. 4, B–D).
Fig. 4.
CD44high type II cells form alveolar-like organoids in 3-D culture. A: schematic showing the culture conditions used for organoid formation. Five thousand CD44high or CD44low type II cells sorted from SpC-CreER/Rosa-mTmG mice were mixed with 100,000 Mlg fibroblast cells and cultured in a 50% Matrigel/collagen mixture inside a 24-well Transwell insert, and 600 μl of culture medium were added outside the Transwell insert. B and C: representative images of GFP+ colonies formed with CD44low (B) or CD44high (C) type II cells after 14 days of culture. Scale bar = 1,000 μm. D: scatterplot showing the sizes of individual GFP+ colonies formed from CD44low or CD44high type II cells. More large colonies were formed with CD44high type II cells. The sizes are presented as areas of the cysts, measured with the ImageJ program using images represented by B and C. Cells from two to four mice were pooled in each experiment, and data are representative of four independent experiments. E–H: immunohistochemistry of sections of GFP+ colonies showed that both CD44low (E and F) and CD44high (G and H) type II cells formed colonies expressing the type II cell marker Sp-C and the type I cell markers T1α (E and G) and HopX (F and H). CD44high type II cells formed colonies arranged into alveolar-like structures with a large lumen inside (*; G and H). In contrast, CD44low type II cells formed colonies that were smaller with no clear identifiable lumen (E and F). I: some cells in the CD44high type II cell-derived colonies maintained CD44 expression after 2 wk of culture. Scale bar = 50 μm for E–I. J: proportions of type II cells (AT2) or type I cells (AT1) relative to all cells in the colonies were scored on the basis of Sp-C and HopX staining and are plotted for CD44low or CD44high cultures. K: CD44low and CD44high cells were cultured in Matrigel mixed with or without HMW HA for 11 days. The sizes of individual colonies formed are plotted for these culture conditions.
The cultures were then processed for sectioning and immunofluorescent antibody-staining analysis. We found that cells were arranged into alveolar-like cysts with a clear lumen only in the large colonies formed by CD44high cells. Sp-C-expressing cuboidal type II cells were located at the periphery of cysts, and flat squamous cells expressing the type I markers T1α and HopX were located in the interior of the cyst wall lining the lumen (Fig. 4, G and H). CD44low cells formed colonies that also consisted of Sp-C-expressing type II cells and type I cells expressing T1α or HopX, but the inside lumen was barely detectable (Fig. 4, E and F). The expression of CD44 was maintained in some cells in CD44high cell-derived colonies after 2 wk of culture (Fig. 4I). The proportions of type II and type I cells, however, were similar in colonies generated from CD44high or CD44low cells (Fig. 4J).
Because the larger volume in the CD44high cell-derived colonies can also be explained by a higher level of liquid secretion, we compared the level of cystic fibrosis transmembrane conductance regulator (CFTR; 17, 19) gene expression in CD44high and CD44low cells. However, the transcript level of CFTR was similar in these two subgroups of cells (Chen Q and Liu Y, unpublished data). To test whether the cultured cells respond to HMW HA, a predominant form of HA present in uninjured lungs (12) and a possible CD44 ligand (12), we cultured CD44high and CD44low cells in Matrigel mixed with HMW HA. Compared with plain Matrigel, both CD44high and CD44low cells formed more large colonies in the presence of HMW HA, but still, more large colonies were formed in the CD44high cell culture than in the CD44low cell culture (Fig. 4K). The differences in colony size of this 3-D culture indicated that CD44high type II cells have a higher progenitor potential compared with their CD44low counterparts, and HMW HA appeared to play a role in stimulating colony growth.
CD44high type II cells are localized in proximity to capillaries.
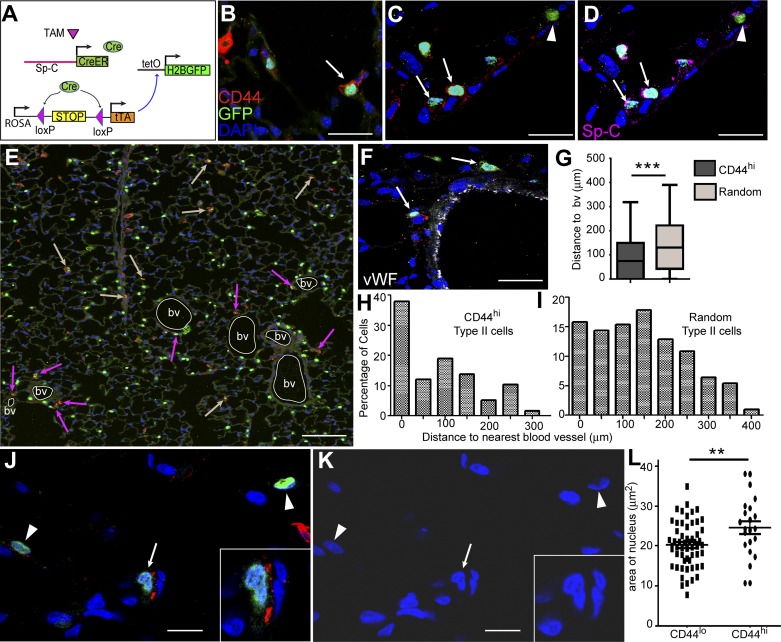
To determine the spatial localization of CD44high type II cells in alveoli, we performed immunohistological analysis using another lineage-tracing line SpC-CreER/Rosa-tTA/tetO-H2B-GFP (Fig. 5A), in which a loxP-flanked stop sequence normally disrupts the expression of tetracycline transactivator (tTA) protein (34) and the subsequent tetO-dependent H2B-GFP expression in cells (33, 34). After tamoxifen injection, Cre was specifically activated in Sp-C+ cells, leading to the excision of the stop sequence, activation of transcription factor tTA, and subsequent expression of nuclear-localized GFP in type II cells. By examining lung sections prepared from this line, we found that GFP+CD44high type II cells were rare and sparsely located in peripheral alveoli, consistent with the FACS results indicating they represent only ~3% of type II cells (Fig. 5B). The CD44high type II cells expressed Sp-C at levels similar to those of the CD44low type II cells (Fig. 5, C and D). Some GFP− cells also showed high CD44 signals; these might represent myeloid cells according to the P28 mouse lung RNAseq data from the LungMap database of the National Heart, Lung, and Blood Institute (https://www.lungmap.net/). Notably, CD44high type II cells appeared to be preferentially located adjacent to the von Willebrand factor (vWF; 32)-labeled blood vessels of the lung (Fig. 5, E and F). By scoring >60 CD44high type II cells and >200 randomly selected type II cells, we found that significantly more CD44high type II cells were located adjacent to blood vessels (Fig. 5, G–I). In addition, the CD44high type II cells had slightly larger nuclei compared with those of CD44low type II cells (Fig. 5, J–L).
Fig. 5.
CD44high type II cells are preferentially located in pericapillary niche. A: schematic showing the labeling strategy for type II cells in SpC-CreER/Rosa-tTA/tetO-H2B-GFP mice. Adult mice were given four doses of tamoxifen (TAM; 0.25 mg/g) 2 wk before lung isolation. Sp-C+ cell-specific Cre then excises the stop codon upstream of tTA, leaving it to activate the tetO promoter, which drives the expression of H2B-GFP in Sp-C+ cells. B: immunohistochemistry analysis of lung sections from these mice showed that only a small fraction of GFP+ cells expressed CD44 at the cell membrane (arrow). C and D: the same lung sections showed staining with GFP and CD44 (C) and GFP and Sp-C (D). The arrows show that GFP+CD44high cells were also positive for Sp-C. Arrowheads indicate cells that were GFP+Sp-C+ type II cells but had no CD44 expression. E: 6 × 6 tiled confocal image showing the distribution of CD44high cells in a larger area of the lung section. Blood vessels (bv) are outlined by white circles, pink arrows show GFP+CD44high cells that are adjacent to bv, and tan arrows indicate those cells that are farther away from bv. F: blood vessels in the lung were labeled with vWF. Arrows show GFP+CD44high cells located adjacent to bv. G–I: GFP+CD44high cells were more frequently detected at perivascular regions of the lung. Sixty-two GFP+CD44high type II cells from five 6 × 6 tiled pictures captured at random locations in lung sections (H) and 204 random GFP+ type II cells in those pictures (I) were analyzed for their distance to the nearest bv; the percentage of the cells vs. the distance to bv is plotted. The mean distance to nearest bv is significantly less for the GFP+CD44high type II cells compared with the random GFP+ type II cells (G); data are presented as Tukey box plot. J–L: areas of nuclei were measured using ImageJ for CD44high (arrows and insets) and CD44low (arrowheads) type II cells and are plotted (L). K is the same image as J but only shows DAPI. **P < 0.01, ***P < 0.001. Scale bar = 30 μm for B, C, D, and F. Scale bar = 150 μm for E. Scale bar = 10 μm for J and K.
Comparing CD44high type II cells with previously identified putative lung stem cells.
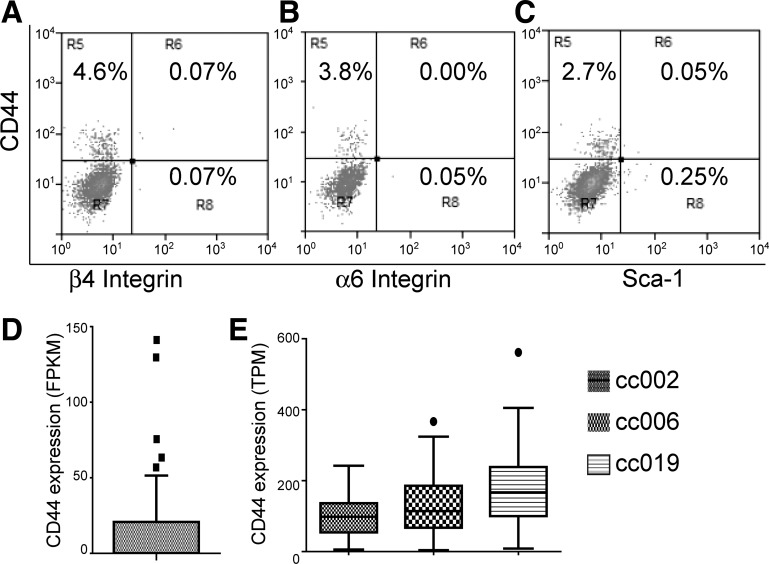
Next, we investigated whether the CD44high type II cells belong to any of the putative stem cell populations previously reported. Using the SpC-CreER/Rosa-Tomato mice, flow cytometry analysis revealed that the Tomato+CD44high type II cells were negative for integrin-α6 and integrin-β4 (Fig. 6, A and B). Therefore the CD44high type II cell population is distinct from the regenerating alveolar cells reported by Chapman et al. (3). Moreover, Tomato+CD44high cells were also negative for Sca-1 (Fig. 6C), which distinguished them from the Sca-1+ bronchioalveolar stem cells (BASC) reported by Kim et al. (15).
Fig. 6.
CD44high type II cells are negative for integrin-α6, integrin-β4, and Sca-1 and can be identified from published single-cell RNAseq analysis data. A–C: type II cells were isolated from SpC-CreER/Rosa-Tomato mice that had been given four doses of tamoxifen (0.25 mg/g) and were subjected to FACS analysis. Tomato-positive cells were gated (not shown) and further analyzed for the expression of CD44, integrin-α6, integrin-β4, and Sca-1. Numbers indicate the percentage of cells in the indicated fraction out of the total gated cells. The data are representative of at least two independent experiments. D: FPKM values of CD44 of 46 lineage-labeled adult mouse type II cells (accession no. GSE52583) are plotted. E: TPM values of CD44 from 84, 47, and 77 type II cells from 3 healthy donors (accession no. GSE84147) are plotted. Data are presented as Tukey box plots.
Recently, several single-cell RNAseq data sets of type II cells have been published (33). We downloaded these data from the Gene Expression Omnibus database and analyzed whether a CD44high type II cell subset can be identified from these studies. In a study by Treutlein et al. (33), 46 lineage-labeled adult mouse type II cells were subjected to single-cell RNAseq (accession no. GSE52583). We analyzed the transcript levels [expressed as fragments per kilobase of transcripts per million mapped read (FPKM; 33)] of CD44 in these cells and found that 5 cells expressed high CD44 levels above the upper Tukey fence (1.5× IQR above the 3rd quartile of the data set) compared with the other 41 cells (Fig. 6D). In another study by Xu et al. (38), 86, 50, and 79 alveolar epithelial cells (97% were type II cells) from 3 normal adult humans were subject to single-cell RNAseq, and we obtained the data using accession no. GSE84147 and further analyzed the expression levels of CD44 [expressed as transcripts per kilobase million (TPM)] in these cells. The seven non-type-II cells (38) were excluded from our analysis on the basis of low Sp-C expression. As shown in Fig. 6E, one cell from donor cc006 and one from donor cc019 expressed high levels of CD44 above the upper Tukey fence. Thus, despite the differences in species and normalization methods of the RNAseq data, these two single-cell RNAseq studies both suggest the presence of a small population of CD44high type II cells.
Aged mice have more CD44high type II cells, but these cells have lower progenitor potentials.
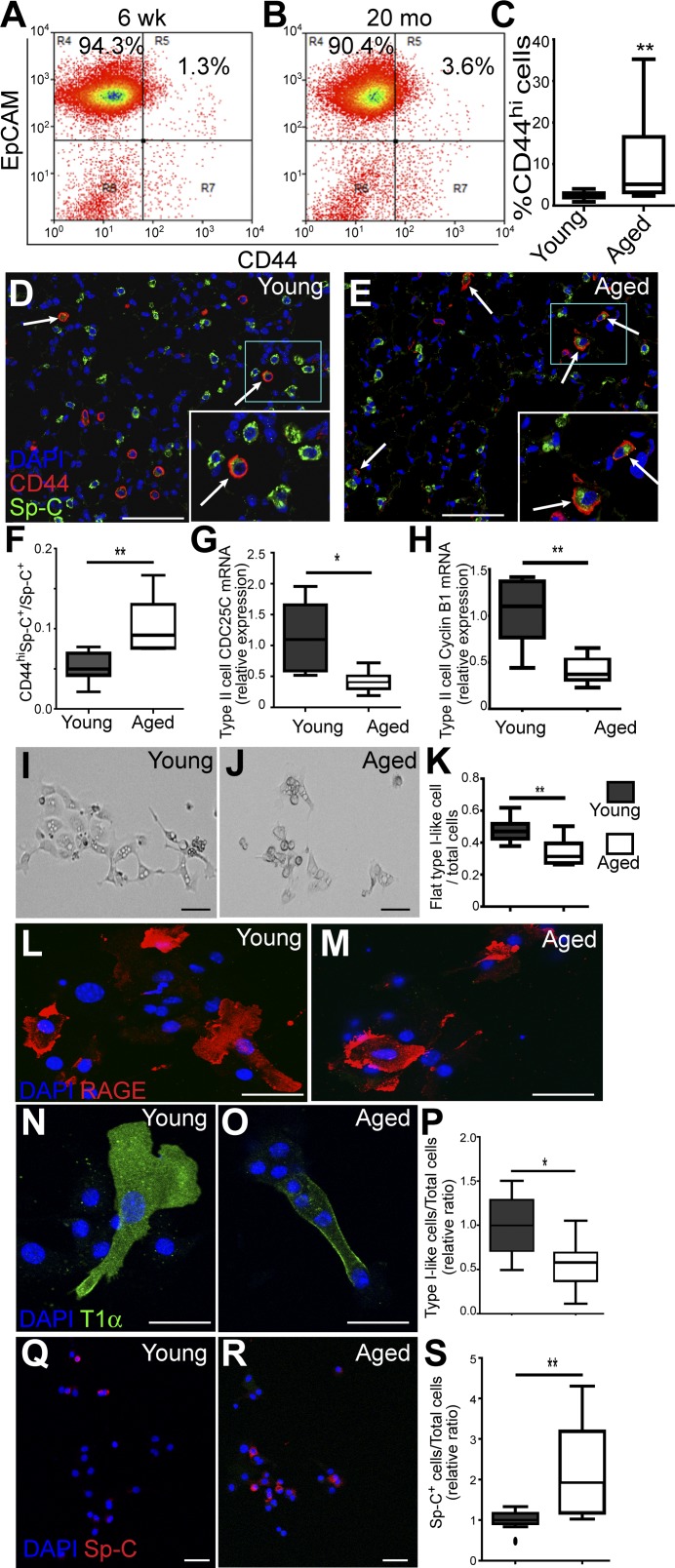
Our data showed that CD44high type II cells exhibit progenitor cell properties and thus may represent a subpopulation important for constitutive alveolar homeostasis. Therefore we next ask whether the CD44high type II cells in lungs change with the aging of the mice. Type II cells were isolated from 6–8-wk-old (young) and 18–24-wk-old (aged) C57BL/6 mice using a type II cell enrichment protocol that gives >90% purity (21). EpCAM was included in FACS analysis to exclude the contaminating cells (such as fibroblast, blood, and endothelial cells) to further increase the purity of type II cells (21). Our FACS analysis showed that the percentage of CD44high type II cells among total type II cells was significantly higher in aged mice compared with young mice (Fig. 7, A–C). To confirm this result, we stained the lung sections prepared from young and aged mice, scored the CD44highSp-C+ cells among the Sp-C+ cells, and found that the proportion of CD44high type II cells among total type II cells was higher in aged mice compared with young mice (Fig. 7, D–F).
Fig. 7.
Aged mice have more CD44high type II cells, but these cells exhibit decreased progenitor properties. A and B: type II cells isolated from 6–8-wk-old (young) or 18–24-mo-old (aged) C57BL/6 mice were analyzed for the presence of EpCAM+CD44high cells. C: percentages of EpCAM+CD44high cells among all the EpCAM+ type II cells are plotted for young and aged mice; n = 11 mice for each age group. D–F: young (D) or aged (E) C57BL/6 mice were subjected to paraffin sectioning and antibody staining, and the percentages of CD44highSp-C+ vs. all Sp-C+ cells were scored and are plotted. Five to eight mice were scored for each age group (F). Arrows in D and E show CD44highSp-C+ cells. Insets in white rectangles are enlarged pictures of blue rectangles. G and H: EpCAM+CD44high cells of the two age groups were isolated by FACS and subjected to qRT-PCR analysis for expression of CDC25C (G) and cyclin B1 (CCNB1; H); n = 7 mice. I–K: EpCAM+CD44high cells from young (I) and aged mice (J) were cultured on gelatin-coated slides for 3 days. Images are representative of similar observations for six to eight mice for each group. The ratios of flat type I-like cells vs. total cells are plotted. For each mouse, 50–200 cells were scored (K). L–O: cultured EpCAM+CD44high cells from young (L and N) and aged (M and O) mice were stained for type I cell markers RAGE (L and M) or T1α (N and O). P: the ratios of cells that expressed either type I cell marker (type I-like cells) vs. total cells were scored; n = 5–7 mice for each group. Q and R: cultured cells from young (Q) and aged (R) mice were stained for type II cell marker Sp-C. S: the ratios of cells that maintained Sp-C expressions were scored; n = 10 mice for each group. Because of the slight variation in culture condition of each independent experiment, P and S are presented as the relative value compared with the average ratios of cells from young mice in the experiment conducted at same time. C, F–H, K, P, and S are Tukey box plots; *P < 0.05, **P < 0.01. Scale bar = 30 μm.
Next, EpCAM+CD44high type II cells of young and aged mice were isolated using FACS sorting and subjected to real-time RT-PCR analysis. Our data showed that the CD44high type II cells of the aged mice expressed significantly lower levels of CDC25C and CCNB1 compared with that of young mice (Fig. 7, G and H), indicating that the aged CD44high type II cells had a lower proliferation rate. Furthermore, when isolated CD44high type II cells of the aged and young mice were cultured for 3 days on gelatin-coated slides, more of the young CD44high type II cells acquired flat type I cell-like morphology compared with these cells from the aged mice (Fig. 7, I–K). We next stained the cultured CD44high type II cells from young or aged mice with type I cell markers RAGE and T1α, as well as type II cell marker Sp-C. We found that a significantly higher proportion of cells from young mice expressed type I cell markers T1α or RAGE compared with those from aged mice (Fig. 7, L–P). In contrast, the ratio of cells that maintained Sp-C expression was higher in cells from aged than from young mice (Fig. 7, Q–S). Thus our data indicate that the CD44high type II cells in aged mice had lower potential for proliferation or transition into type I cells compared with young mice.
DISCUSSION
Previous lineage-tracing studies using type II cell-specific promoters showed that without injury, the percentage of cells lineage-labeled with the type II marker Sp-C stayed constant over a long period (up to 48 wk; 1, 4), indicating that type II cells are replenished by self-renewal of Sp-C+ cells during the steady-state condition, rather than being derived from other cell types (1, 4). Here, we found that in lungs in the absence of injury, Sp-C+CD44high type II cells showed distinct progenitor cell properties including higher proliferation compared with other type II cells. These results suggested that these cells are the specific subpopulation of type II cells involved in constitutively renewing the alveolar epithelium. In addition, CD44high type II cells also exhibited higher potential to differentiate into type I cells in culture. In 2-D culture, they appeared to be more elongated and spread to a larger area, which is a phenotype indicating more mature type I cells (39). They also had higher potential for differentiating into cells expressing type I cell markers. In a 3-D culture system, the CD44high type II cells were more likely to form alveolar-like colonies composed of both type II and type I cells. These experiments indicated that the CD44high type II cells might also play a role in giving rise to type I cells during homeostatic maintenance of the lung.
Significantly, in aged mice there were more CD44high type II cells, but these cells showed decreased progenitor potential in that they expressed lower levels of cell proliferation genes CDC25C and CCNB1 and also had lower potential for transition into type I-like cells. The increased number of CD44high type II cells might represent a mechanism of compensation for their loss of function. This phenomenon is consistent with the finding of aging hematopoietic stem cells (HSC): the numbers of several groups of phenotypically defined HSCs increase with aging, but these aged HSCs showed decreased regenerative potential (8). The dynamic changes of the number and progenitor properties of the CD44high type II cells with aging further supported that they are a subgroup of progenitor type II cells for the steady-state maintenance of alveolar epithelial cells.
We found that CD44high type II cells were preferentially located in alveoli close to vWF-expressing pulmonary capillaries. This preferential location of CD44high type II cells as well as their slightly larger nuclei resemble the characteristics of founder type II cells in the “hot spots” of alveolar epithelial renewal foci described by Desai et al. during steady-state homeostasis (4). This location suggests that the components associated with microvessels may serve as an important niche for these cells. Although CD44 is a cell surface glycoprotein essential for homing of stem cells to their niches (13, 16, 41), it is not clear whether these cells express a higher CD44 level permanently or only transiently overexpress CD44 when engaged in self-renewal and type I cell transition. Nevertheless, we have found by 3-D culture that some cells in the CD44high type II cell-derived colonies maintained CD44 expression after 2 wk of culture. Thus higher levels of CD44 expression can be maintained in cells after multiple cell cycles. A CD44 promoter-directed lineage-tracing mouse line will be helpful to further study the properties of CD44high cells and the relationship of CD44 and the niche.
We found that HMW HA had an effect in increasing the colony formation capacities of both CD44high and CD44low cells, and its effect in CD44high cells was more significant. These results suggest that HA acts as a ligand for CD44 in vitro. However, CD44 does not appear to be required for the proliferation of type II cells in vivo, as lungs from CD44null mice showed a type II cell BrdU incorporation rate similar to that of wild-type controls (Chen Q, Kumar VS, and Liu Y, unpublished data). This result indicates that CD44 might be redundant for some of the type II cell progenitor functions, and therefore it may serve as a surface marker for the progenitors rather than play an essential role for the cell proliferation per se.
By analyzing recently published single-cell RNAseq results for mouse and human type II cells (33, 38), we found that there is indeed a small subset of type II cells expressing higher levels of CD44 than the rest; thus our observations are consistent with these data. Other than type II cells, several populations of alveolar epithelial cells were shown to possess progenitor properties: Sca-1+ BASC (15), Sca-1+ type II cells (21), and alveolar stem cells expressing transformation-related protein-63 (Trp63), keratin 5, and integrin-α6β4 (18) (3, 35, 42). We found that the CD44high type II cells were negative for expression of Sca-1, integrin-α6, or integrin-β4 and thus are different from these previously reported cells.
In conclusion, we identified a novel subgroup of CD44high type II cells that are preferentially located near blood vessels. These cells exhibit a distinct progenitor marker and behavior compared with other type II cells in lungs in the basal state, and their progenitor properties decrease after aging. Therefore it is likely that the CD44high type II cells play a role in maintaining alveolar homeostasis.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant 105947 (Y. Liu) and Training Grant 7829-23 (J. Finn).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Q.C., V.S.K., J.F., D.J., J.L., Y.-Y.Z., and Y.L. conceived and designed research; Q.C., V.S.K., J.F., and Y.L. performed experiments; Q.C., V.S.K., J.F., and Y.L. analyzed data; Q.C., V.S.K., J.F., D.J., J.L., Y.-Y.Z., and Y.L. interpreted results of experiments; Q.C., V.S.K., and Y.L. prepared figures; Q.C., V.S.K., and Y.L. drafted manuscript; Q.C. and Y.L. edited and revised manuscript; Q.C., V.S.K., J.F., D.J., J.L., Y.-Y.Z., and Y.L. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Chonghui Cheng (Baylor College of Medicine) for discussion and providing reagents. We thank Dr. Peng Jiang (Northwestern University) and Dr. Mark Maienschein-Cline (Center for Research Informatics, University of Illinois at Chicago) for assistance in analyzing the single-cell RNAseq data.
REFERENCES
- 1.Barkauskas CE, Michael J, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123: 3025–3036, 2013. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest 121: 1064–1074, 2011. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, Vu TH. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 121: 2855–2862, 2011. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507: 190–194, 2014. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobbs LG. Isolation and culture of alveolar type II cells. Am J Physiol Lung Cell Mol Physiol 258: L134–L147, 1990. [DOI] [PubMed] [Google Scholar]
- 6.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Renewal of alveolar epithelium in the rat following exposure to NO2. Am J Pathol 70: 175–198, 1973. [PMC free article] [PubMed] [Google Scholar]
- 7.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp Mol Pathol 22: 142–150, 1975. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- 8.Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat Rev Immunol 13: 376–389, 2013. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- 9.Goodell MA, Nguyen H, Shroyer N. Somatic stem cell heterogeneity: diversity in the blood, skin and intestinal stem cell compartments. Nat Rev Mol Cell Biol 16: 299–309, 2015. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gracz AD, Fuller MK, Wang F, Li L, Stelzner M, Dunn JC, Martin MG, Magness ST. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells 31: 2024–2030, 2013. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu YC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med 20: 847–856, 2014. doi: 10.1038/nm.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol Rev 91: 221–264, 2011. doi: 10.1152/physrev.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med 12: 1167–1174, 2006. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 14.Kasper M, Günthert U, Dall P, Kayser K, Schuh D, Haroske G, Müller M. Distinct expression patterns of CD44 isoforms during human lung development and in pulmonary fibrosis. Am J Respir Cell Mol Biol 13: 648–656, 1995. doi: 10.1165/ajrcmb.13.6.7576702. [DOI] [PubMed] [Google Scholar]
- 15.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121: 823–835, 2005. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 16.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med 12: 1175–1180, 2006. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 17.Kreda SM, Davis CW, Rose MC. CFTR, mucins, and mucus obstruction in cystic fibrosis. Cold Spring Harb Perspect Med 2: a009589, 2012. doi: 10.1101/cshperspect.a009589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, Wang Y, Lim B, Chow VT, Crum CP, Xian W, McKeon F. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147: 525–538, 2011. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazrak A, Jurkuvenaite A, Chen L, Keeling KM, Collawn JF, Bedwell DM, Matalon S. Enhancement of alveolar epithelial sodium channel activity with decreased cystic fibrosis transmembrane conductance regulator expression in mouse lung. Am J Physiol Lung Cell Mol Physiol 301: L557–L567, 2011. doi: 10.1152/ajplung.00094.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Kim J, Gludish D, Roach RR, Saunders AH, Barrios J, Woo AJ, Chen H, Conner DA, Fujiwara Y, Stripp BR, Kim CF. Surfactant protein-C chromatin-bound green fluorescence protein reporter mice reveal heterogeneity of surfactant protein C-expressing lung cells. Am J Respir Cell Mol Biol 48: 288–298, 2013. doi: 10.1165/rcmb.2011-0403OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Kumar VS, Zhang W, Rehman J, Malik AB. Activation of type II cells into regenerative stem cell antigen-1(+) cells during alveolar repair. Am J Respir Cell Mol Biol 53: 113–124, 2015. doi: 10.1165/rcmb.2013-0497OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Sadikot RT, Adami GR, Kalinichenko VV, Pendyala S, Natarajan V, Zhao YY, Malik AB. FoxM1 mediates the progenitor function of type II epithelial cells in repairing alveolar injury induced by Pseudomonas aeruginosa. J Exp Med 208: 1473–1484, 2011. doi: 10.1084/jem.20102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason RJ. Biology of alveolar type II cells. Respirology 11, Suppl: S12–S15, 2006. doi: 10.1111/j.1440-1843.2006.00800.x. [DOI] [PubMed] [Google Scholar]
- 24.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell 18: 8–23, 2010. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Protin U, Schweighoffer T, Jochum W, Hilberg F. CD44-deficient mice develop normally with changes in subpopulations and recirculation of lymphocyte subsets. J Immunol 163: 4917–4923, 1999. [PubMed] [Google Scholar]
- 26.Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4: 525–534, 2009. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy R, Buckley S, Doerken M, Barsky L, Weinberg K, Anderson KD, Warburton D, Driscoll B. Isolation of a putative progenitor subpopulation of alveolar epithelial type 2 cells. Am J Physiol Lung Cell Mol Physiol 286: L658–L667, 2004. doi: 10.1152/ajplung.00159.2003. [DOI] [PubMed] [Google Scholar]
- 28.Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, Noble PW, Hogan BL. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA 108: E1475–E1483, 2011. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneeberger EE. Alveolar type I cells. In: The Lung: Scientific Foundations(2nd ed.), edited by Crystal RG, West JB, Weibel ER, and Barnes PJ. Philadelphia, PA: Lippincott Williams & Wilkins, 1997, p. 535–542. [Google Scholar]
- 30.Stripp BR. Hierarchical organization of lung progenitor cells: is there an adult lung tissue stem cell? Proc Am Thorac Soc 5: 695–698, 2008. doi: 10.1513/pats.200801-011AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szabo AZ, Fong S, Yue L, Zhang K, Strachan LR, Scalapino K, Mancianti ML, Ghadially R. The CD44+ ALDH+ population of human keratinocytes is enriched for epidermal stem cells with long-term repopulating ability. Stem Cells 31: 786–799, 2013. doi: 10.1002/stem.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torisu T, Torisu K, Lee IH, Liu J, Malide D, Combs CA, Wu XS, Rovira II, Fergusson MM, Weigert R, Connelly PS, Daniels MP, Komatsu M, Cao L, Finkel T. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med 19: 1281–1287, 2013. doi: 10.1038/nm.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509: 371–375, 2014. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science 303: 359–363, 2004. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, Matthay MA, Rock JR, Chapman HA. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517: 621–625, 2015. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer 14: 468–480, 2014. doi: 10.1038/nrc3744. [DOI] [PubMed] [Google Scholar]
- 37.Weibel ER. What makes a good lung? Swiss Med Wkly 139: 375–386, 2009. smw-12270. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, Wikenheiser-Brokamp KA, Perl AT, Funari VA, Gokey JJ, Stripp BR, Whitsett JA. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1: e90558, 2016. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Hernandez BJ, Martinez Alanis D, Narvaez del Pilar O, Vila-Ellis L, Akiyama H, Evans SE, Ostrin EJ, Chen J. The development and plasticity of alveolar type 1 cells. Development 143: 54–65, 2016. doi: 10.1242/dev.130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao YY, Liu Y, Stan RV, Fan L, Gu Y, Dalton N, Chu PH, Peterson K, Ross J Jr, Chien KR. Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc Natl Acad Sci USA 99: 11375–11380, 2002. doi: 10.1073/pnas.172360799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 11: 254–267, 2011. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 42.Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, Crum CP, Xian W, McKeon F. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517: 616–620, 2015. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]