Figure 4.
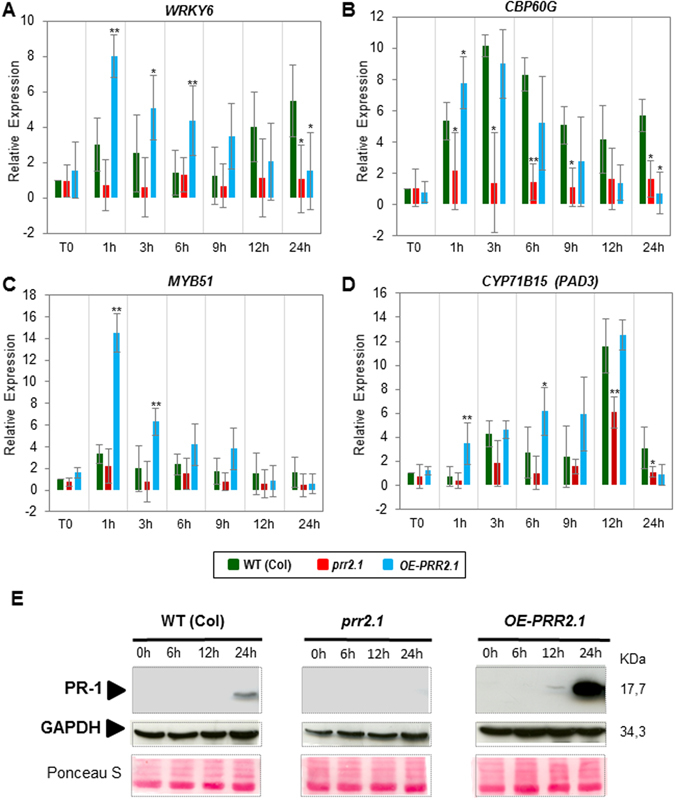
Expression analysis of defence marker genes and detection of PR1 protein in prr2 knock-down mutants and in transgenic line overexpressing PRR2. (A,B,C and D) Analyses of WRKY6 (A), CBP60G (B), MYB51 (C), CYP71B15/PAD3 (D) marker genes in different prr2 genetic backgrounds. Leaves of 4-week-old Arabidopsis WT (Col), mutant prr2.1, and OE-PRR2.1 were inoculated with 5 × 107 cfu.mL−1 of Pst DC3000 and harvested at 0, 1, 3, 6, 9, 12 and 24 hours post-inoculation (hpi). The fold changes relative to the mock treatment were determined by RT-Q PCR. The values are means ± standard deviation of three independent experiments. Asterisks (*) above histograms indicate significant changes of PRR2 gene expression in these different genetic backgrounds compared to WT (Col) (student t-test with p-value < 0.05 (*) or p-value 0.01 (**)). (E) Detection of PR1 by immunoblot experiment in WT plants and prr2 genotypes. PR1 accumulation was detected in leaves of 3-week-old plants at 0, 6, 12, 24 hpi after spraying with Pst DC3000 (2 × 108 cfu.mL−1). Equal loading was confirmed by immunoblot detection of GAPDH and Ponceau S staining of the membrane (middle and lower panels). The illustrated blot is representative of three biological replicates.
