Abstract
Background/Aims:
Accurate and rapid laboratory diagnosis of Clostridium difficile infections (CDI) remains a significant challenge. A two-step algorithm for detection of toxigenic C. difficile in stool based on initial screening for glutamate dehydrogenase assay followed by confirmation by toxin A+B detection using an enzyme immunoassay (EIA) or molecular assay has been proposed. We aimed to evaluate the C. difficile Quik Chek Complete® (QCC-EIA) versus the GeneXpert® C. difficile polymerase chain reaction (PCR) assay in this two-step algorithm.
Materials and Methods:
Two hundred and ten liquid stool samples obtained between June 2014 and June 2015 from patients suspected of CDI were tested by the QCC-EIA and GeneXpert PCR assay. The GeneXpert assay was used as the reference standard to calculate the QCC-EIA sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
Results:
Of the 210 stool samples tested, 43 (20.5%) were positive by QCC-EIA, while 31 (14.8%) were positive by GeneXpert assay. The sensitivity and specificity of the QCC-EIA were found to be 100 and 93%, respectively; the PPV and NPV were 72 and 100%, respectively. The binary toxin was detected in 12 (38.7%) and tcdC gene deletion in 3 (9.6%).
Conclusions:
The low specificity of QCC-EIA makes it less reliable as a confirmatory test for CDI diagnosis. This test may be used as a screening test in a two-step algorithm when combined with a molecular assay or another confirmatory test.
Keywords: Clostridium difficile, enzyme immunoassay, GeneXpert PCR assay
INTRODUCTION
Clostridium difficile is a major etiological agent of healthcare associated infections worldwide. C. difficile infection is associated with significant patient mortality, morbidity, prolonged hospitalization, and increased healthcare cost. Rapid and accurate identification of toxigenic C. difficile is important for early commencement of appropriate management and implementation of infection control measures with resultant improvement in patient outcomes.[1] Laboratory tests available for C. difficile diagnosis include stool culture, enzyme immunoassays (EIAs) for toxin detection, glutamate dehydrogenase (GDH) assay for antigen detection, cell culture cytotoxicity neutralization assay (CCNA), and molecular assays. However, the choice of test is influenced by cost, sensitivity and specificity, turn-around-time, and ease of performance in the diagnostic laboratory. For example, although both stool culture and CCNA have high sensitivity, they are more expensive and have longer turn-around-time. In contrast, EIA for detection of toxins A and B is a simple inexpensive test but low sensitivities have been reported depending on the type of kit, study population, and gold standard test used.[2,3] The Society for Healthcare Epidemiology of America and Infectious Disease Society of America guidelines indicate that EIAs should no longer be considered as adequate stand-alone tests for the diagnosis of C. difficile infection.[4] Tests like cell CCNAs and toxigenic culture are unattractive confirmatory tests in the diagnostic laboratory because they are complex and tedious. Molecular assays represent better alternative as confirmatory tests as they are easier to perform and have higher specificity and sensitivity.[5,6]
Recently, a two-step testing algorithm for detecting toxigenic C. difficile directly from the stool of symptomatic patients was proposed.[7] This algorithm is based on a GDH EIA for initial screening, followed by toxin A+B detection using EIA or molecular assay for confirmation. The TechLab C. difficile Quik Chek Complete® (QCC-EIA) (Techlab, Blacksburg, VA, USA) is a rapid EIA for the simultaneous detection of C. difficile GDH antigen and toxins A/B, while the GeneXpert C. difficile polymerase chain reaction (PCR) assay (Cepheid, CA, USA) is a real-time PCR test that detects the toxin B gene (tcdB), the binary toxin gene (cdt), and the tcdC gene deletion at nt 117 in C. difficile.[6] In this study, we have carried out an evaluation of the utilization of the QCC-EIA versus the GeneXpert C. difficile PCR assay in this two-step algorithm.
MATERIALS AND METHODS
Stool samples
A total of 210 stool samples obtained from patients presenting with diarrhea and clinically suspected of C. difficile infection were tested. These were samples submitted to the microbiology laboratory at King Khalid University Hospital, Riyadh, Kingdom of Saudi Arabia from June 2014 to June 2015 as part of routine clinical investigations. Presence of diarrhea was an entry criterion. All stool samples were liquid stool and no formed stool was accepted for the study. The detection of C. difficile toxin was carried out using QCC-EIA and GeneXpert C. difficile PCR assay. Samples were stored and refrigerated at 2–8°C.
Quik chek complete-enzyme immunoassay
The detection of GDH antigen and/or toxins by QCC-EIA was carried out in accordance with the manufacturers' instructions, including the use of appropriate controls, as specified in the package insert. Briefly, ~25 ml of stool sample was added to a tube containing the diluent and conjugate and the mixture was transferred to the device sample well. After incubation for 15 min at room temperature, the wash buffer followed by the substrate were added to the reaction window. The results were read after 10 min. The GDH antigen and/or toxins were reported as positive if a clear visible band was seen on the antigen and/or toxin side of the device display window, respectively, as per manufacturer guidelines.
GeneXpert C. difficile PCR assay: A sterile cotton tipped swab was dipped into the watery stool sample and placed in sample reagent and vortexed at high speed for 10 seconds at room temperature. All liquid from the sample reagent was transferred into the “S” chamber of GeneXpert C. difficile cartridge as stipulated in the package insert. Reagent one was added to chamber one and reagent two to chamber two of the test cartridge. The cartridge barcode was scanned and the cartridge was placed in the GeneXpert instrument for the PCR run. The resulting data were interpreted as positive, negative, or invalid as per manufacturer recommendations.
Statistical analysis
Data were analyzed using Statistical Package for the Social Sciences (SPSS) version 18.0 (SPSS Inc., Chicago, Illinois, USA). GeneXpert C. difficile PCR assay was used as the reference standard to calculate the assay sensitivity, specificity, PPV and NPV.
RESULTS
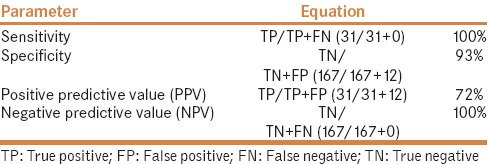
Majority of the stool samples (n = 110; 52.4%) were from males. The mean age (±SD) of the patient population was 42 (±24.3) years. Most of the specimens were from inpatients (n = 171; 81.4%) mainly from the medical ward (n = 61). Of the 210 stool samples tested, 43 (20.5%) were positive by QCC-EIA, while 31 (14.8%) were positive by GeneXpert C. difficile PCR assay. Among those positive by GeneXpert C. difficile PCR assay, 100% (n = 31) were positive for toxin B, 38.7% (n = 12) for binary toxin, and 9.6% (n = 3) for tcdC gene deletion. All 31 samples positive by GeneXpert C. difficile PCR assay were also positive by QCC-EIA and were considered as true positives [Table 1]. There were 12 false positive findings but no false negatives [Table 1]. Based on these findings, the sensitivity of QCC-EIA was 100%, whereas the specificity was 93% [Table 2]. In addition, the PPV was 72%, while the NPV was 100%. Table 2 shows the calculation of the parameters.
Table 1.
Comparison of QCC-EIA with GeneXpert C. difficile PCR assay
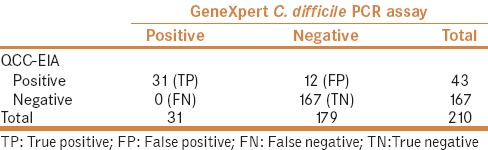
Table 2.
Sensitivity, specificity, and positive and negative predictive value of QCC-EIA
DISCUSSION
We have evaluated the performance of QCC-EIA test versus GeneXpert C. difficile PCR assay for the detection of C. difficile toxin in the stool of patients suspected of CDI. Our findings indicate that although the QCC-EIA has high sensitivity for the C. difficile toxin detection in stool samples, it lacks specificity. In contrast to our findings, previous reports suggest that C. difficile toxin EIA lacks sensitivity but shows better specificity compared to molecular test and the cell culture cytotoxin neutralization assay for detection of C. difficile toxin.[8,9,10] However, emerging data now indicate that sensitivity and specificity of the EIA assay might be related to the C. difficile ribotype present in the stool sample being tested.[11] We speculate that this might explain the differences in the sensitivity and specificity of the QCC-EIA assay observed in our study compared to other reports. The virulent C. difficile strain BI/NAP-1/027 carries a mutation in the tcdC genetic locus resulting in a truncated protein tcdC, which leads to hyper-production of toxins A and B.[12,13] In our study, the tcdC gene deletion was identified in three stool samples and to our knowledge, this is the first positive report of tcdC deletion and binary toxin in stool samples collected from Saudi patients. This finding is suggestive of the presence of C. difficile strain BI/NAP-1/027 in our setting. This is of concern as C. difficile BI/NAP1/027 is associated with high-level fluoroquinolone resistance and outbreaks. These findings indicate the need for urgent work to determine the prevalent ribotypes associated with C. difficile infection in Saudi Arabia. Available report on C. difficile ribotypes in Saudi Arabia revealed that 027, BT1, V, and A2 have been identified in retail food.[14]
Another finding in our study is the impact of test performance on the predictive value of the QCC-EIA. Although the QCC-EIA has a high NPV that correlates with absence of disease in patients suspected of CDI, the PPV is only 72%. This data implies that a C. difficile QCC-EIA positive result would require another confirmatory test to obtain a definitive diagnosis of CDI. Some laboratories have utilized the C. difficile toxin EIA in a two-step procedure for diagnosis of C. difficile infection.[5,10] This algorithm involves C. difficile EIA as a screening test and a PCR assay as confirmatory test. While the two-step algorithm may be cost saving, it is more laborious and prolongs the turn-around-time for reporting final results when compared to a stand-alone PCR for C. difficile toxin.[10] In view of superior performance characteristic and a shorter turn-around-time, many clinical laboratories have now implemented the molecular assay. The American Society for Microbiology guidance document for laboratory detection of C. difficile toxin recommends the use of molecular assay as a stand-alone test, while the C. difficile toxin EIA results should be confirmed with either a toxigenic culture or a molecular assay.[15]
CONCLUSION
In conclusion, the QCC-EIA is useful as a screening test for the detection of C. difficile toxin A + B in stool samples. Due to its low specificity, a second confirmatory test is required, which may include a molecular assay. The GeneXpert C. difficile PCR assay demonstrates a superior accuracy of toxin B detection, has a shorter turn-around-time with simplicity of sample preparation, and it can detect gene targets associated with hyper-virulent strains of C. difficile. Utilization of the GeneXpert C. difficile PCR in the laboratory diagnosis of CDI will be invaluable in promoting early initiation of management interventions, enhancing patient outcomes and reducing the incidence of healthcare associated infections.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Martin JS, Monaghan TM, Wilcox MH. Clostridium difficile infection: Epidemiology, diagnosis and understanding transmission. Nat Rev Gastroenterol Hepatol. 2016;13:206–16. doi: 10.1038/nrgastro.2016.25. [DOI] [PubMed] [Google Scholar]
- 2.Alcala L, Sanchez-Cambronero L, Catalan MP, Sanchez-Somolinos M, Pelaez MT, Marin M, et al. Comparison of three commercial methods for rapid detection of Clostridium difficile toxins A and B from fecal specimens. J Clin Microbiol. 2008;46:3833–5. doi: 10.1128/JCM.01060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sloan LM, Duresko BJ, Gustafson DR, Rosenblatt JE. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J Clin Microbiol. 2008;46:1996–2001. doi: 10.1128/JCM.00032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–55. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 5.Novak-Weekley SM, Marlowe EM, Miller JM, Cumpio J, Nomura JH, Vance PH, et al. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010;48:889–93. doi: 10.1128/JCM.01801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, et al. Impact of strain type on detection of toxigenic Clostridium difficile: Comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010;48:3719–24. doi: 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478–98. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 8.Gilligan PH. Optimizing the laboratory diagnosis of Clostridium difficile infection. Clin Lab Med. 2015;35:299–312. doi: 10.1016/j.cll.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Jamal W, Pauline EM, Rotimi VO. Comparative performance of the GeneXpert C. difficile PCR assay and C. diff Quik Chek Complete kit assay for detection of Clostridium difficile antigen and toxins in symptomatic community. onset infections. Int J Infect Dis. 2014;29:244–8. doi: 10.1016/j.ijid.2014.10.025. [DOI] [PubMed] [Google Scholar]
- 10.Tenover FC, Baron EJ, Peterson LR, Persing DH. Laboratory diagnosis of Clostridium difficile infection can molecular amplification methods move us out of uncertainty? J Mol Diagn. 2011;13:573–82. doi: 10.1016/j.jmoldx.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajabally N, Kullin B, Ebrahim K, Brock T, Weintraub A, Whitelaw A, et al. A comparison of Clostridium difficile diagnostic methods for identification of local strains in a South African Centre. J Med Microbiol. 2016 doi: 10.1099/jmm.0.000231. [DOI] [PubMed] [Google Scholar]
- 12.Kociolek LK, Gerding DN. Clinical utility of laboratory detection of Clostridium difficile strain BI/NAP1/027. J Clin Microbiol. 2016;54:19–24. doi: 10.1128/JCM.02340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curry SR, Marsh JW, Muto CA, O'Leary MM, Pasculle AW, Harrison LH. tcdC genotypes associated with severe tcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol. 2007;45:215–21. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alqumber MA. Clostridium difficile in retail baskets, trolleys, conveyor belts, and plastic bags in Saudi Arabia. Saudi Med J. 2014;35:1274–7. [PMC free article] [PubMed] [Google Scholar]
- 15.American Society of Microbiology: A Practical Guidance Document for the Laboratory Detection of Toxigenic Clostridium difficile. [Last accessed on 2017 Feb 07]. Available from: https://www.asm.org/images/pdf/Clinical/clostridiumdifficile9-21.pdf .


