SUMMARY
Zika viruses (ZIKVs) are circulating in parts of the world endemic for other flavivirus infections. Some cross-reactive antibodies (Abs) elicited by prior flavivirus exposures can bind to ZIKV and enhance infection of Fc receptor-bearing cells. Here, we measured ZIKV binding of 54 murine monoclonal Abs (mAbs) elicited by exposure with Dengue virus and West Nile virus antigens. We found that 8 of 54 mAbs recognized the envelope protein of ZIKV in conventional binding assays. These 8 cross-reactive mAbs have different specificities; most recognize the DI/II region of the envelope protein but one mAb recognized the DIII lateral ridge of the envelope protein. Interestingly, only 3 of these cross-reactive mAbs were able to enhance ZIKV infection in vitro, and enhancing potential was not strictly correlated with relative binding ability. These data suggest that the ability of flavivirus Abs to enhance ZIKV is dependent on multiple factors.
Keywords: zika virus, antibody, antibody mediated enhancement, flavivirus
INTRODUCTION
Zika virus (ZIKV) is a mosquito-transmitted flavivirus first isolated from a Rhesus monkey in the Zika forest in Uganda in 1947 (Dick et al., 1952). ZIKV outbreaks have been historically confined to areas of Africa and Asia (Fauci and Morens, 2016) and most viral infections are associated with mild symptoms such as fever and headache (Bearcroft, 1956; Simpson, 1964). Over the past year, ZIKVs rapidly spread throughout the Americas and other parts of the world (Hennessey et al., 2016). Unlike past outbreaks, the current ZIKV epidemic is of great concern since it has been associated with microcephaly in fetuses and newborns and Guillain-Barre syndrome in adults (Cao-Lormeau et al., 2016; Carteaux et al., 2016; Oehler et al., 2014). It is unclear why neurological symptoms have not been linked to ZIKV infections prior to 2013. It is possible that this is due to underreporting, or that contemporary ZIKV strains have acquired genetic changes that alter disease progression. Alternatively, the host populations most affected by the current ZIKV outbreak may be more pre-disposed to develop ZIKV-elicited neurological disease compared to host populations affected by earlier ZIKV outbreaks.
How can different host populations have different susceptibility to ZIKV-elicited severe disease? One possibility is that host populations affected by past and current ZIKV outbreaks have different immune histories. ZIKVs are currently circulating in many parts of the world that are endemic for other flaviviruses, and prior viral exposures can actually enhance disease caused by infections with antigenically distinct flaviviruses. For example, antibodies (Abs) elicited against one serotype of DENV can enhance disease caused by a different serotype of DENV through a process called Ab-dependent enhancement (ADE) (Halstead and O’Rourke, 1977). ADE can occur when immune complexes, formed between cross-reactive Abs and virus, are targeted to Fc receptor-bearing cells that are permissive for viral replication (Halstead, 2015; Schmid et al., 2014; Whitehead et al., 2007). Several studies have shown that DENV-elicited Abs can enhance ZIKV replication in vitro and in vivo (Bardina et al., 2017; Charles and Christofferson, 2016; Dejnirattisai et al., 2016; Paul et al., 2016; Stettler et al., 2016; Zhao et al., 2016).
Here, we completed a series of studies to better define the characteristics of Abs that are capable of enhancing ZIKV replication in vitro. We characterized 54 murine monoclonal Abs (mAbs) that were previously elicited by DENV and West Nile Virus (WNV) antigens. We measured relative Ab binding and ability to enhance infection of a contemporary strain of ZIKV in vitro. Our data indicate that Abs of different specificities and relative binding abilities can enhance ZIKV infection in vitro.
Results
mAbs elicited by DENV and WNV bind to ZIKV
We obtained a large panel of mouse mAbs elicited by DENV and WNV antigens from BEI Resources (Supplemental table 1), most of which were created by Michael Diamond (Washington University) and colleagues (Brien et al., 2010; Gentry et al., 1982; Henchal et al., 1982; Oliphant et al., 2005; Oliphant et al., 2006; Shrestha et al., 2010; Sukupolvi-Petty et al., 2013). These mAbs include 20 mAbs elicited by DENV serotype 1 (DENV-1), 2 mAbs elicited by DENV-2, 14 mAbs elicited by DENV-3, 11 mAbs elicited by DENV-4, and 7 mAbs elicited by WNV. This panel of mAbs includes 33 IgG1, 14 IgG2a, 5 IgG2b, and 2 IgG2c. All of the mAbs bind the envelope (E) protein of the respective eliciting antigen, and most bind to domains I/II (DI/II) (n=24 mAbs) or DIII (n=26 mAbs) of the E protein (Fig 1).
Fig 1.
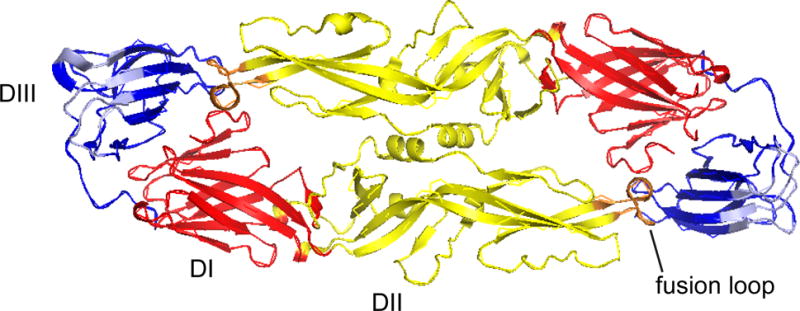
ZIKV E protein structure. Domain 1 (D1), red; D11, yellow; DIII, blue; fusion loop, orange; and DIII lateral ridge, light blue are highlighted on the previously published ZIKV E protein structure (PDB: 5JHM) (Dai et al., 2016) using Pymol.
First, we used conventional ELISAs to measure binding of each mAb to a contemporary strain of ZIKV isolated from Puerto Rico in 2015 (PRVABC59) (Fig 2). We considered a mAb to be ZIKV-reactive if the ZIKV ELISA signal was >2x the ELISA signal of the mAb against a no-antigen control and of an irrelevant influenza virus-specific mAb (170-3C12) against PRVABC59 ZIKV in three independent experiments. Six of 49 DENV-elicited and 2 of 7 WNV-elicited mAbs were considered ZIKV-reactive using these criteria. We also included a ZIKV-specific mAb (ZIKV-0402166) as a control. Interestingly, several of the DENV- or WNV-elicited mAbs bound to ZIKV more strongly than the ZIKV-0402166 mAb.
Fig 2.
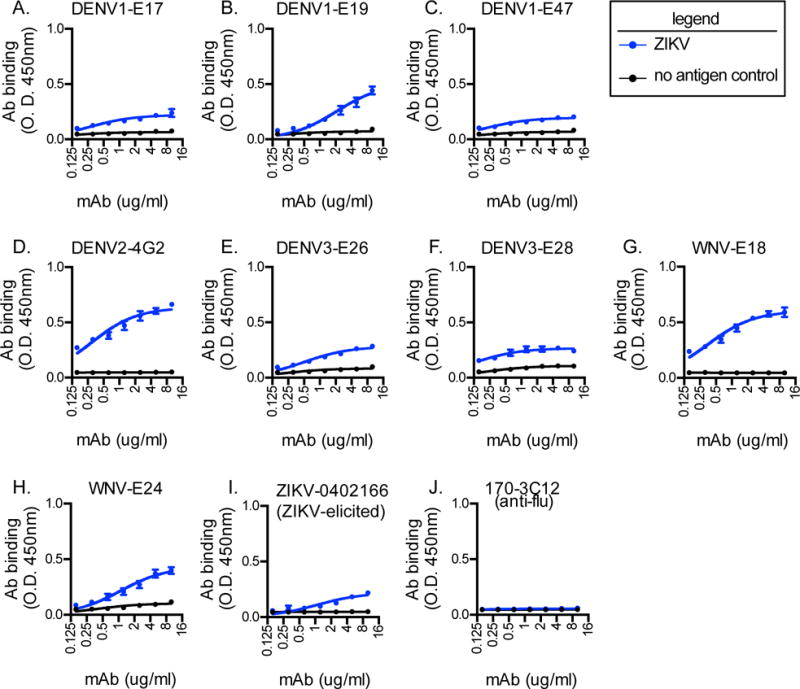
DENV- and WNV-elicited mAbs bind to ZIKV PRVABC59. We tested a panel of mAbs for binding to ZIKV PRVABC59 (blue line) or a no-antigen control (black line) by ELISA. Samples were run in triplicate and data are shown as mean +/− SD. Data shown are representative of 3 independent experiments. We included the ZIKV-0402166 mAb (ZIKV-elicited) as a positive control and 170-3C12 mAb (influenza virus-elicited) as a negative control. Binding curves were calculated using one-site specific binding. We tested 54 mAbs and shown are the mAbs where the ZIKV ELISA signal was >2x the ELISA signal of the mAb against a no-antigen control and of the irrelevant influenza virus-specific mAb (170-3C12).
Next, we used flow cytometry to measure binding of these 8 cross-reactive mAbs to ZIKV-infected Vero cells. All of the Abs that bound to ZIKV by ELISA also bound to infected cells (Figure 3). Three of the mAbs (4G2, WNV-E18, and WNV-E24) showed relatively high variance in binding to infected Veros (see wider range of fluorescence intensities in Figure 3). This binding pattern suggests that the epitopes of these mAbs are variably present 24 hours post-infection. This may be related to viral breathing of immature virions, heterogeneity in the viral population, or different stages of maturity of the viral particle within individual cells (Diamond and Pierson, 2015). It is also possible that this might relate to heterogeneity in the mAb preparations or differential reactivity of the anti-mouse IgG secondary Ab used in these experiments.
Fig 3.
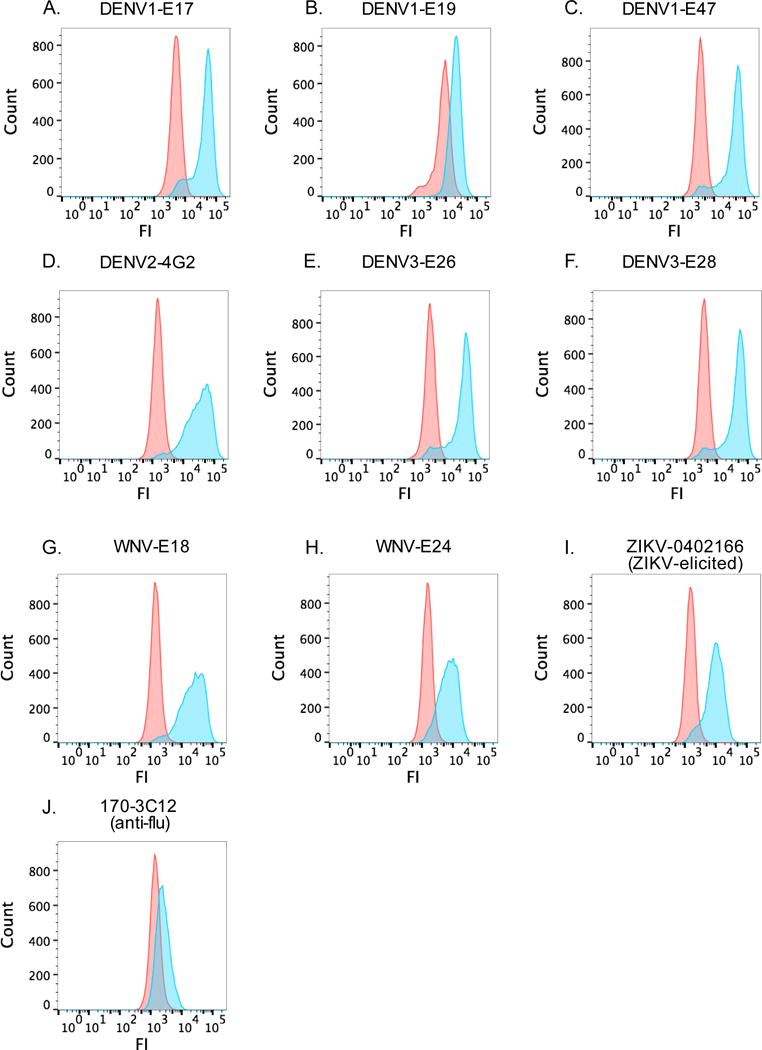
DENV- and WNV-elicited mAbs bind to ZIKV PRVABC59-infected cells.
We tested the mAbs identified in Figure 2 for binding to infected Vero cells by flow cytometry. Vero cells were infected with ZIKV PRVABC59 at moi=5. The next day, the cells were fixed and permeabilized and 15ug/ml of each mAb was added, followed by the addition of a FITC-conjugated anti-mouse IgG Ab. We included the ZIKV-0402166 mAb (ZIKV-elicited) as a positive control and 170-3C12 mAb (influenza virus-elicited) as a negative control. Data are representative of 2 independent experiments. FI, fluorescent intensity.
DENV- and WNV-elicited mAbs enhance ZIKV infection in vitro
Previous studies suggest that some DENV-elicited mAbs can inhibit ZIKV replication (Barba-Spaeth et al., 2016; Priyamvada et al., 2016), while other DENV-elicited mAbs enhance ZIKV infection of Fc receptor-bearing cells (Charles and Christofferson, 2016; Dejnirattisai et al., 2016; Paul et al., 2016; Priyamvada et al., 2016; Stettler et al., 2016; Zhao et al., 2016). To determine whether the 8 cross-reactive mAbs in our study were able to neutralize ZIKV in vitro, we completed focus reduction neutralization tests (FRNTs) which assess the ability of Abs to block infection of Vero cells, which do not express Fc receptors. None of the 8 cross-reactive mAbs were able to neutralize ZIKV infection of Vero cells at concentrations up to 10ug/ml (Fig 3a). In contrast, as a positive control, we found that sera collected from ZIKV-infected mice were able to neutralize ZIKV in these assays (Fig 4a).
Fig 4.
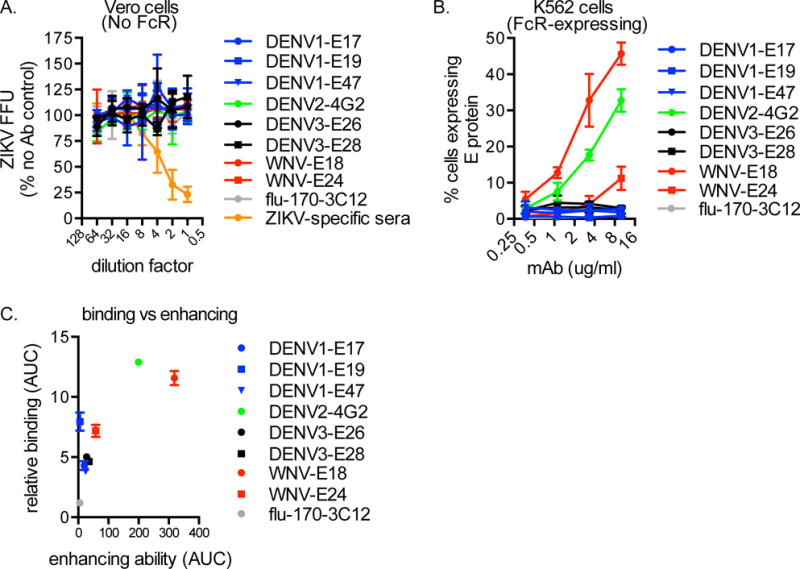
DENV- and WNV-elicited mAbs enhance ZIKV PRVABC59 in vitro. The mAbs identified in Figure 2 were tested for neutralization of ZIKV PRVABC59 by FRNTs in Vero cells. mAbs (initial concentration 10 ug/ml) or anti-ZIKV polyclonal mouse sera (positive control; initial dilution 1:40) were incubated with virus at 37C for 1h, then the inoculum was adsorbed to cells for 1h. After 40h, intracellular E protein expression was determined using the 4G2 mAb, and the number of foci per well were counted manually. Shown are data (mean +/− SD) compiled from 3 independent experiments. FFU, focus forming unit. (B) Next, mAbs were tested for enhancement of ZIKV PRVABC59 infection of K562 cells. mAbs were incubated with virus at 37C for 1h, then added to cells. 3 days later, cells were collected and intracellular E protein expression was determined using flow cytometry. Data (mean +/− SD) are representative of 3 independent experiments. (C) Enhancement ability was calculated as the area under the curve (AUC) of each mAb in (B) and plotted against relative binding (mean +/− SD as determined by AUC; Figure 2).
We next completed similar experiments in K562 cells, which are a human erythroleukemic cell line that express Fc receptors. We infected K562 cells +/− each mAb and then used flow cytometry to measure ZIKV E protein expression 3 days later. Infection in the presence of both WNV-elicited mAbs, as well as 4G2, increased the percentage of K562 cells expressing ZIKV E protein (Fig 4b). The 3 mAbs that enhanced infection of K562 cells are IgG2a isotype, while the non-enhancing mAbs are mostly IgG1 (and one IgG2b). In mice, IgG2a mAbs have been associated with ADE (Halstead, 2003); however, in this in vitro system utilizing a human cell line, the predominance of IgG2a enhancing mAbs is likely due to the higher affinity of murine IgG2a than IgG1 or IgG2b for human Fc receptors (Lubeck et al., 1985). There was not a strict correlation between mAb relative binding and enhancement (Fig 4c). Notably, the 3 mAbs that enhanced infection of K562 cells are the same mAbs that displayed higher variance in binding to infected Vero cells (Fig 3).
DISCUSSION
Most studies of ADE have focused on DENVs. There is substantial evidence that ADE contributes to severe disease during secondary DENV infections in humans (Diamond and Pierson, 2015; Halstead, 2003). In this context, ADE is thought to occur when a prior exposure to one DENV serotype elicits Abs that are able to bind to, but not effectively neutralize, DENV of a different serotype. Ab-DENV immune complexes can be internalized by Fc receptor-bearing cells such as monocytes and macrophages, which are permissive for DENV replication, leading to increased viral replication (Diamond and Pierson, 2015; Goncalvez et al., 2007; Zellweger et al., 2010). ADE disease severity observed in vivo may also be due to increased cytokine release and vascular permeability (Balsitis et al., 2010; Beatty et al., 2015). Recent studies suggest that ADE is associated with IgGs that have high affinity for activating Fc receptors (Wang et al., 2017).
Several recent studies have demonstrated that DENV-specific mAbs or DENV-elicited polyclonal sera can enhance ZIKV infection in vitro (Charles and Christofferson, 2016; Dejnirattisai et al., 2016; Paul et al., 2016; Stettler et al., 2016; Zhao et al., 2016) and a recent study demonstrated that DENV and WNV-elicited Abs can enhance ZIKV infection in vivo (Bardina et al., 2017). In the current study, we tested ZIKV binding and enhancing potential of a large panel of murine flavivirus mAbs and we identified mAbs elicited by DENV and WNV antigens that are capable of enhancing ZIKV infection in vitro. All enhancing mAbs were of the IgG2a subclass and displayed similar binding patterns as determined by flow cytometry.
The ZIKV E protein is closely related to DENV E proteins (54–57% amino acid conservation) and the WNV E protein (53% amino acid conservation) (Barba-Spaeth et al., 2016). One of the 3 enhancing mAbs (4G2) in our studies was elicited by DENV. DENVs are prevalent in Brazil and other areas of the world that have been greatly affected by the current ZIKV outbreak (Messina et al., 2014). It is interesting that 2 of 3 enhancing mAbs identified in our studies were elicited by WNV. Zhao et al. also observed in vitro enhancement of ZIKV infection by a mAb elicited by WNV (Zhao et al., 2016). WNV has been reported in northeastern regions of Brazil (Vieira et al., 2015) that have had high levels of ZIKV-associated microcephaly (Brasil et al., 2016; Hazin et al., 2016; Mlakar et al., 2016). The WNV-specific enhancing Abs we identified bind to the DII fusion loop (WNV-E18) and the DIII lateral ridge (WNV-E24) of the E protein (Oliphant et al., 2005; Oliphant et al., 2006), while the DENV-elicited mAb (4G2) also binds to the DII fusion loop (Crill and Chang, 2004; Stiasny et al., 2006) (Fig. 1). Paul et al. identified two human DENV-elicited mAbs that bind to the fusion loop and enhance ZIKV infection in vitro (Paul et al., 2016). As a class, fusion loop Abs are highly cross-reactive since this epitope is well conserved among flaviviruses (Crill and Chang, 2004; Dejnirattisai et al., 2016). DENV-specific mAbs with different specificities have been shown to enhance infection of DENV (Dejnirattisai et al., 2016; Halstead, 2003) although studies suggest that fusion loop Abs are more potent at inducing ADE than Abs specific for other epitopes (Dejnirattisai et al., 2016).
Taken together, our data suggest that Abs elicited by different flavivirus exposures and Abs of different specificities can enhance ZIKV infection in vitro. It remains to be seen if DENV and WNV-elicited Abs of different specificities contribute to disease in vivo. Ultimately, a better understanding of how prior flavivirus infections influence ZIKV infections will be important for designing new ZIKV vaccines and for understanding the unusual patterns of disease associated with the current ZIKV outbreak.
EXPERIMENTAL PROCEDURES
Cells and virus
PRVABC59, a 2015 ZIKV isolate from Puerto Rico, was obtained from BEI Resources (NR-50240) and grown in Vero cells. For ELISA stocks, virus was concentrated by ultracentrifugation. K562 cells, a human erythroleukemic cell line, were a gift from J. Riley (U. Penn).
Antibodies
DENV- and WNV-elicited mAbs were obtained from BEI Resources (see supplemental table), except for 4G2, which was obtained either from BEI Resources, or as ascites fluid (EMD Millipore, Billerica, MA) and then purified over NAb Protein G Spin Column (ThermoScientific, Rockland, IL). Two clones obtained from BEI Resources (DENV3-E11 and DENV3-E2) bound non-specifically by ELISA or to uninfected cells and were omitted from analyses. An influenza virus-specific mAb (170-3C12) was also obtained from BEI Resources. The ZIKV-elicited mAb (ZIKV-0402166) was purchased from Aalto Bioreagents (Dublin, Ireland).
ELISAs
Immulon 4HBX 96-well plates (ThermoScientific, Rochester, NY) were coated overnight at 4C with concentrated PRVABC59 diluted in PBS, or PBS alone. Plates were blocked with 3% BSA in PBS for 2h at room temperature, then washed 5x in deionized water. mAbs were serially diluted in 1% BSA in PBS, added to the plates, and incubated for 2h at room temperature. Plates were washed 5x then HRP-conjugated anti-mouse IgG (MP Biomedicals, Santa Ana, CA) diluted 1:2500 in 1% BSA was added and incubated for 1h at room temperature. Plates were developed with 3,3′,5,5′-Tetramethylbenzidine (TMB) SureBlue (KPL, Gaithersburg, MD) and read with a SpectraMax 190 plate reader (Molecular Devices, Sunnyvale, CA). Binding curves were plotted in GraphPad Prism (San Diego, CA) using one-site specific binding. mAbs were classified as positive if binding of the mAb at a fixed concentration was >2x both binding of the mAb to no-antigen control and binding of negative control mAb 170-3C12 to PRVABC59 in 3 independent ELISAs.
Flow cytometry binding assay
Vero cells were infected with PRVABC59 (moi 5), or left uninfected. The next day, cells were collected and washed in 1% BSA in PBS then in PBS alone. Cells were fixed and permeabilized with BD Perm/Fix (BD Cytofix/Cytoperm Plus Kit, BD Biosciences, San Diego, CA), then stained with mAbs diluted to 15ug/ml in 1x Perm/Wash (BD Cytofix/Cytoperm Plus Kit, BD Biosciences, San Diego, CA). Cells were then stained with FITC-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) and FITC signal evaluated by flow cytometer (LSRII, BD Biosciences, San Diego, CA). Data were analyzed using FlowJo (Ashland, OR).
Focus reduction neutralization test (FRNT)
Vero cells were plated at 2.5 × 104 cells/well in 96 well plates (BD, Corning, NY) the day before the assay. mAbs and sera were serially diluted in serum-free media and mixed with a constant amount of virus, then incubated at 37C for 1h. Ab/virus mixtures were added to washed cells, then incubated at 37C for 1h. Inocula were removed and cells were washed with serum-free media, and overlaid with 1.25% Avicel RC-591 NF (FMC BioPolymer, Philadelphia, PA) in plaque assay media. After incubation at 37C for 40h, the overlay was removed and cells were fixed with 4% paraformaldehyde (1h, 4C) and permeabilized with 0.5% triton-X (7 min, room temperature). Wells were blocked with 5% BSA in TBS-T (Bio-Rad, Hercules, CA), which was used for subsequent dilutions, for 1h at room temperature. Cells were stained with 4G2 (0.4ug/ml) for 1h at room temperature, then HRP-conjugated anti-mouse IgG (MP Biomedicals, Santa Ana, CA) diluted 1:2000 was added for 1h at room temperature. Wells were developed with TMB TrueBlue (KPL, Gaithersburg, MD) for 2–5h and dried overnight. Images were collected on an ImmunoSpot (Cellular Technology Ltd., Shaker Heights, OH) and foci were counted manually.
Enhancement assay
mAbs were serially diluted in serum-free media and mixed with a constant amount of virus, then incubated at 37C for 1h. K562 cells were added to mAb/virus mixture to a final moi of 0.5, then incubated for 2 days at 37C. Cells were washed in 1% BSA in PBS, incubated with anti-CD16/CD32 (BD Biosciences, San Diego, CA), then fixed and permeabilized (BD Cytofix/Cytoperm Plus, BD Biosciences, San Diego, CA). Cells were stained with anti-DENV 3-E26 (NR-15527, BEI Resources) directly labeled with AF647 using Alexa Fluor 647 Protein Labeling Kit (Molecular Probes, Eugene, OR) and data were collected by flow cytometer (LSRII, BD Biosciences, San Diego, CA). Data were analyzed using FlowJo (Ashland, OR).
Sequence comparisons
Amino acid sequences for ZIKV PRVABC59 (KU501215.1), DENV-1 (NC_001477), DENV-2 (NC_001474), DENV-3 (NC_001475), DENV-4 (NC_002640), and WNV (NC_001563) E proteins were aligned using protein BLAST. Different epitopes were highlighted on the ZIKV E protein structure (PDB ID: 5JHM) (Dai et al., 2016) using PyMol. For this, DIII lateral ridge residues were obtained from (Zhao et al., 2016).
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (U19AI057229, SEH; T32AI055428, EW) and institutional funds from the University of Pennsylvania (SEH). Scott E. Hensley, Ph.D. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes one table.
AUTHOR CONTRIBUTIONS
EW designed experiments, completed all experiments, analyzed data, and wrote the paper.
SEH designed experiments, supervised all experiments, analyzed data and wrote the paper.
References
- Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, Harris E. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS pathogens. 2010;6:e1000790. doi: 10.1371/journal.ppat.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Loriere E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, England P, Stiasny K, Mongkolsapaya J, Heinz FX, Screaton GR, Rey FA. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, Stramer SL, Garcia-Sastre A, Krammer F, Lim JK. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017 doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearcroft WG. Zika virus infection experimentally induced in a human volunteer. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1956;50:442–448. [PubMed] [Google Scholar]
- Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med. 2015;7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- Brasil P, Pereira JP, Jr, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baiao AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. Zika Virus Infection in Pregnant Women in Rio de Janeiro. The New England journal of medicine. 2016;375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, Diamond MS. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. Journal of virology. 2010;84:10630–10643. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra JC, Despres P, Fournier E, Mallet HP, Musso D, Fontanet A, Neil J, Ghawche F. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteaux G, Maquart M, Bedet A, Contou D, Brugieres P, Fourati S, Cleret de Langavant L, de Broucker T, Brun-Buisson C, Leparc-Goffart I, Mekontso Dessap A. Zika Virus Associated with Meningoencephalitis. The New England journal of medicine. 2016;374:1595–1596. doi: 10.1056/NEJMc1602964. [DOI] [PubMed] [Google Scholar]
- Charles AS, Christofferson RC. Utility of a Dengue-Derived Monoclonal Antibody to Enhance Zika Infection In Vitro. PLoS currents. 2016;8 doi: 10.1371/currents.outbreaks.4ab8bc87c945eb41cd8a49e127082620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WD, Chang GJ. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. Journal of virology. 2004;78:13975–13986. doi: 10.1128/JVI.78.24.13975-13986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Song J, Lu X, Deng YQ, Musyoki AM, Cheng H, Zhang Y, Yuan Y, Song H, Haywood J, Xiao H, Yan J, Shi Y, Qin CF, Qi J, Gao GF. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell host & microbe. 2016;19:696–704. doi: 10.1016/j.chom.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, Mongkolsapaya J, Screaton GR. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nature immunology. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Pierson TC. Molecular Insight into Dengue Virus Pathogenesis and Its Implications for Disease Control. Cell. 2015;162:488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- Fauci AS, Morens DM. Zika Virus in the Americas–Yet Another Arbovirus Threat. The New England journal of medicine. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- Gentry MK, Henchal EA, McCown JM, Brandt WE, Dalrymple JM. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am J Trop Med Hyg. 1982;31:548–555. doi: 10.4269/ajtmh.1982.31.548. [DOI] [PubMed] [Google Scholar]
- Goncalvez AP, Engle RE, St Claire M, Purcell RH, Lai CJ. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9422–9427. doi: 10.1073/pnas.0703498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Pathogenesis of Dengue: Dawn of a New Era. F1000Res. 2015;4 doi: 10.12688/f1000research.7024.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB, O’Rourke EJ. Antibody-enhanced dengue virus infection in primate leukocytes. Nature. 1977;265:739–741. doi: 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- Hazin AN, Poretti A, Turchi Martelli CM, Huisman TA, Microcephaly Epidemic Research, G. Di Cavalcanti Souza Cruz D, Tenorio M, van der Linden A, Pena LJ, Brito C, Gil LH, de Barros Miranda-Filho D, Marques ET, Alves JG. Computed Tomographic Findings in Microcephaly Associated with Zika Virus. The New England journal of medicine. 2016;374:2193–2195. doi: 10.1056/NEJMc1603617. [DOI] [PubMed] [Google Scholar]
- Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- Hennessey M, Fischer M, Staples JE. Zika Virus Spreads to New Areas - Region of the Americas, May 2015-January 2016. MMWR. Morbidity and mortality weekly report. 2016;65:55–58. doi: 10.15585/mmwr.mm6503e1. [DOI] [PubMed] [Google Scholar]
- Lubeck MD, Steplewski Z, Baglia F, Klein MH, Dorrington KJ, Koprowski H. The interaction of murine IgG subclass proteins with human monocyte Fc receptors. Journal of immunology. 1985;135:1299–1304. [PubMed] [Google Scholar]
- Messina JP, Brady OJ, Scott TW, Zou C, Pigott DM, Duda KA, Bhatt S, Katzelnick L, Howes RE, Battle KE, Simmons CP, Hay SI. Global spread of dengue virus types: mapping the 70 year history. Trends in microbiology. 2014;22:138–146. doi: 10.1016/j.tim.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T. Zika Virus Associated with Microcephaly. The New England journal of medicine. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. Zika virus infection complicated by Guillain-Barre syndrome–case report, French Polynesia, December 2013. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2014:19. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nature medicine. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. Journal of virology. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul LM, Carlin ER, Jenkins MM, Tan AL, Barcellona CM, Nicholson CO, Michael SF, Isern S. Dengue virus antibodies enhance Zika virus infection. Clin Transl Immunology. 2016;5:e117. doi: 10.1038/cti.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MA, Diamond MS, Harris E. Dendritic cells in dengue virus infection: targets of virus replication and mediators of immunity. Frontiers in immunology. 2014;5:647. doi: 10.3389/fimmu.2014.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O’Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS pathogens. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson DI. Zika Virus Infection in Man. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1964;58:335–338. [PubMed] [Google Scholar]
- Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, Foglierini M, Pedotti M, Simonelli L, Dowall S, Atkinson B, Percivalle E, Simmons CP, Varani L, Blum J, Baldanti F, Cameroni E, Hewson R, Harris E, Lanzavecchia A, Sallusto F, Corti D. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- Stiasny K, Kiermayr S, Holzmann H, Heinz FX. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. Journal of virology. 2006;80:9557–9568. doi: 10.1128/JVI.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. Journal of virology. 2013;87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira MA, Romano AP, Borba AS, Silva EV, Chiang JO, Eulalio KD, Azevedo RS, Rodrigues SG, Almeida-Neto WS, Vasconcelos PF. West Nile Virus Encephalitis: The First Human Case Recorded in Brazil. Am J Trop Med Hyg. 2015;93:377–379. doi: 10.4269/ajtmh.15-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TT, Sewatanon J, Memoli MJ, Wrammert J, Bournazos S, Bhaumik SK, Pinsky BA, Chokephaibulkit K, Onlamoon N, Pattanapanyasat K, Taubenberger JK, Ahmed R, Ravetch JV. IgG antibodies to dengue enhanced for FcgammaRIIIA binding determine disease severity. Science. 2017;355:395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat Rev Microbiol. 2007;5:518–528. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- Zellweger RM, Prestwood TR, Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell host & microbe. 2010;7:128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, Fremont DH. Structural Basis of Zika Virus-Specific Antibody Protection. Cell. 2016;166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


