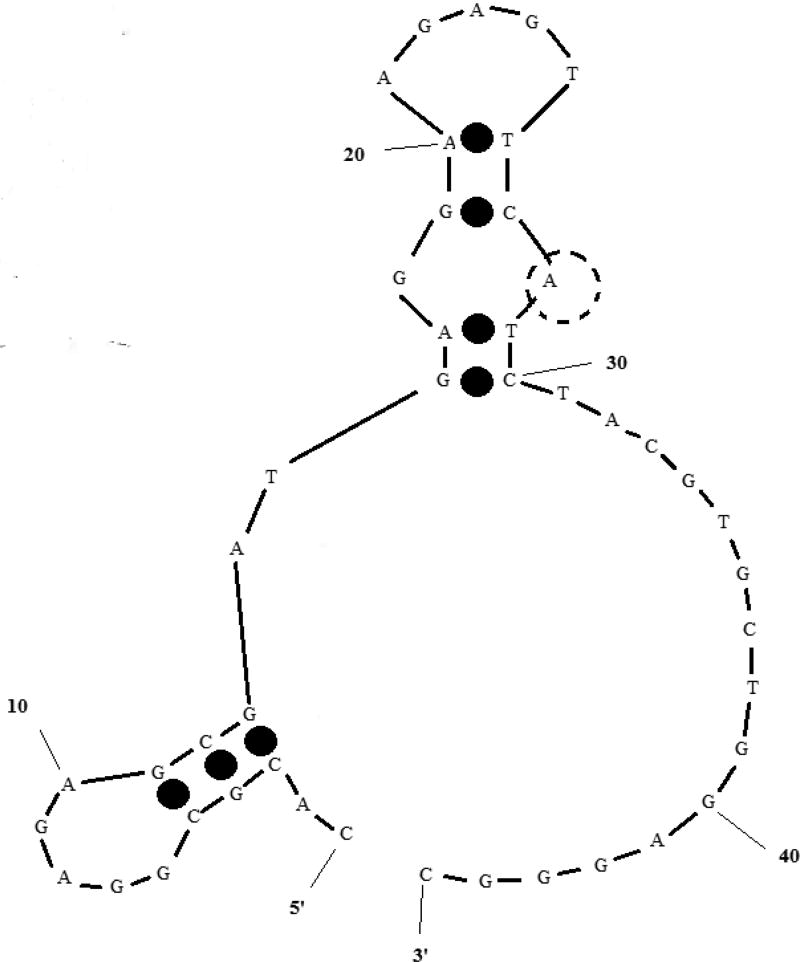
FIGURE 2. Most stable secondary structure of exo probe targeting K. pneumoniae.
ΔG = −3.62 kcal mol−1; the transparent dotted circle (position 28) marks the nucleotide position replaced by THF and eventually cleaved by exonuclease III. RPA salt conditions − 14 mM Mg2+, 100 mM K+ – were used for secondary structure prediction.