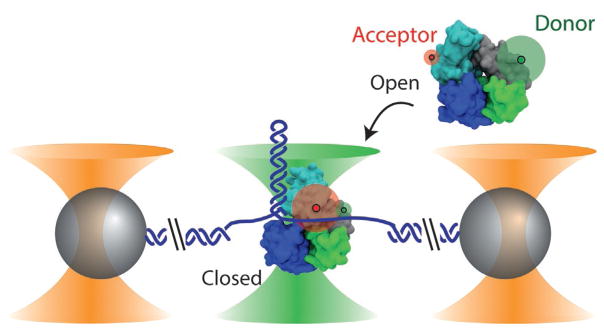
Fig. 1.
Combined high-resolution optical tweezers and confocal microscope. Dual optical traps (outer cones) hold polystyrene microspheres (spheres) tethered by a DNA construct (here a DNA hairpin), while a confocal microscope (middle cone) detects fluorescence from a single molecule. In this example, the conformational and unwinding dynamics of E. coli UvrD helicase are investigated. UvrD helicase exists in two conformational states—“open” (shown in the free protein) and “closed” (shown in the bound protein)—that are differentiated by smFRET between a donor–acceptor pair labeling the protein (green and red disks, respectively). The proteins in this figure were prepared with VMD (Humphrey, Dalke, & Schulten, 1996) from PDB entries 2IS2 and 3LFU. Figure reproduced from Comstock, M. J., Whitley, K. D., Jia, H., Sokoloski, J., Lohman, T. M., Ha, T., & Chemla, Y. R. (2015). Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science, 348(6232), 352–354 with permission from AAAS.