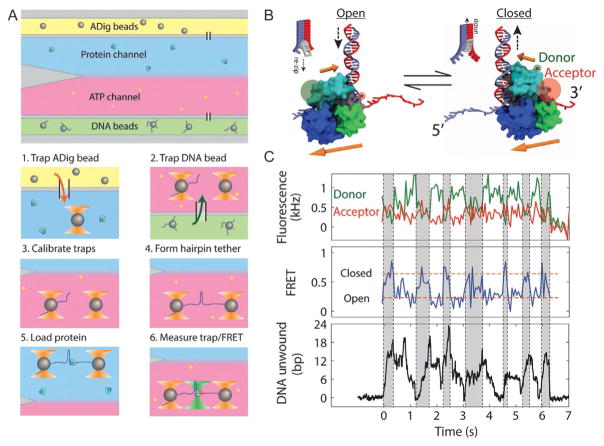
Fig. 9.
Simultaneous measurement of UvrD conformation and activity. (A) Steps for in situ nucleoprotein complex assembly. The flow chamber consists of three channels. The top (yellow) and bottom (green) channels contain antidigoxigenin (ADig) and DNA-coated streptavidin (DNA) beads, respectively, and the central measurement channel consists of two parallel laminar flow streams containing 10 nM protein (blue) and 10 μM ATP (red). During an experiment, we carry out the following steps in sequence: (1) trap an ADig bead dispensed out of the top capillary, (2) move to the bottom capillary and trap a DNA bead, (3) calibrate the traps, (4) form a tether by bringing the beads into contact and pulling apart (and optionally taking a F–x curve), (5) move to the protein stream and load UvrD by incubating for 15 s, and (6) move to the ATP stream, turning on the fluorescence excitation. (B) Model of UvrD conformational switching, in which the closed (open) state corresponds to translocation into (away from) the DNA fork, on opposite strands of the hairpin. (C) Simultaneous measurement of donor (green, top panel) and acceptor (red) fluorescence intensity, FRET efficiency (middle panel), and hairpin base pairs unwound (bottom panel) during UvrD unwinding. A correlation is seen between the “closed,” high FRET state and hairpin unwinding (gray-shaded time intervals) and “open,” low FRET state and hairpin rezipping (unshaded time intervals). Figure adapted from Comstock, M. J., Whitley, K. D., Jia, H., Sokoloski, J., Lohman, T. M., Ha, T., & Chemla, Y. R. (2015). Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science, 348(6232), 352–354 with permission from AAAS.