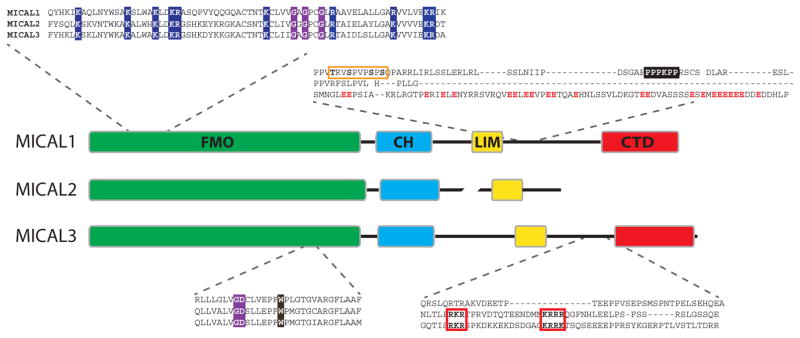
Figure 3. Domains in MICAL proteins.
The domain organization of mammalian MICAL1–3 is schematically represented. Each domain is shown in a different color, conserved along the text: green for FMO, light blue for CH, yellow for LIM and red for CTD. The length of each domain is proportional to the number of amino acids in it, so do linkers, unless braked. Inserts show sequence details of mouse MICAL1–3 (NP_612188.1, NP_001180234.1 and NP_001257404.1, respectively). The conserved basic residues in the N-terminal region of the FMO domain are depicted with blue background and the GxGxxG and GD motifs with purple background. The conserved W is shown over dark brown background. Part of the connecting sequence between LIM and CTD domains is highlighted in the upper part, indicating a tandem of phosphorylation sites on MICAL1 (orange box) and the PPPKPP motif (black background), and the enrichment of acidic residues on MICAL3 (red letters). In the lower panel, the bipartite nuclear localization signal present in MICAL2 and 3 is shown with red boxes and bold letters.