Figure 1.
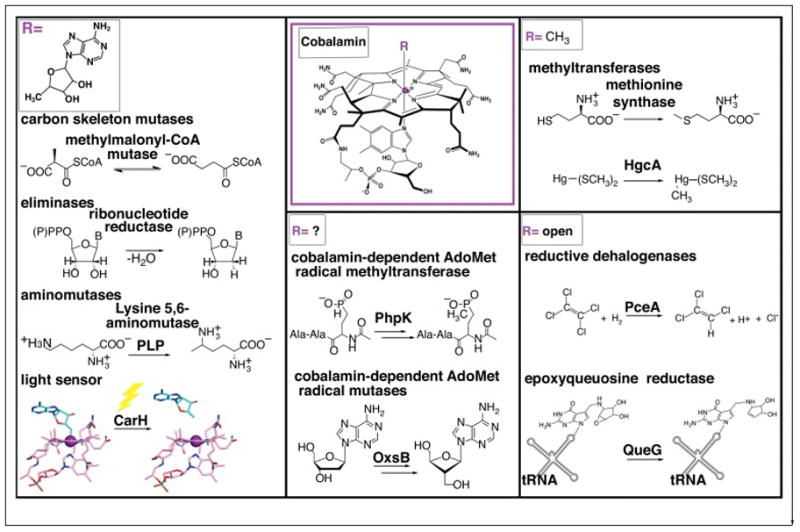
Classes of Cbl-dependent proteins as defined by the nature of the upper ligand. AdoCbl-dependent enzymes include carbon skeleton mutases, eliminases, and aminomutases. The latter of which also requires pyridoxyl phosphate (PLP). Recently, AdoCbl has been shown to be involved in light-dependent gene regulation of the carotenoid (Car) biosynthetic operon via the transcriptional regulator CarH, adding a new function for this flavor of Cbl cofactor. MeCbl-dependent methyl transferases include methionine synthase, which catalyzes the transfer of a methyl cation to homocysteine and the mercury methylase HgcA, which is thought to catalyze transfer of a methyl anion to a Hg2+-bis(thiolate) compound (represented as Hg-S(CH3)2), the most prevalent form of Hg2+ in methylmercury producers [17]. A third class of Cbl-dependent enzymes, which includes the reductive dehalogenase PceA and the queuosine biosynthetic enzyme QueG do not have an upper axial ligand and are denoted as “open”-Cbl-dependent enzymes. The final class is made up of enzymes that appear to require the machinery for both S-adenosylmethionine (AdoMet) radical chemistry and a Cbl cofactor. Examples include phosphinate methylase PhpK and oxetanocin-A biosynthetic enzyme OxsB. The role(s) of Cbl in this enzyme family is currently under investigation.
