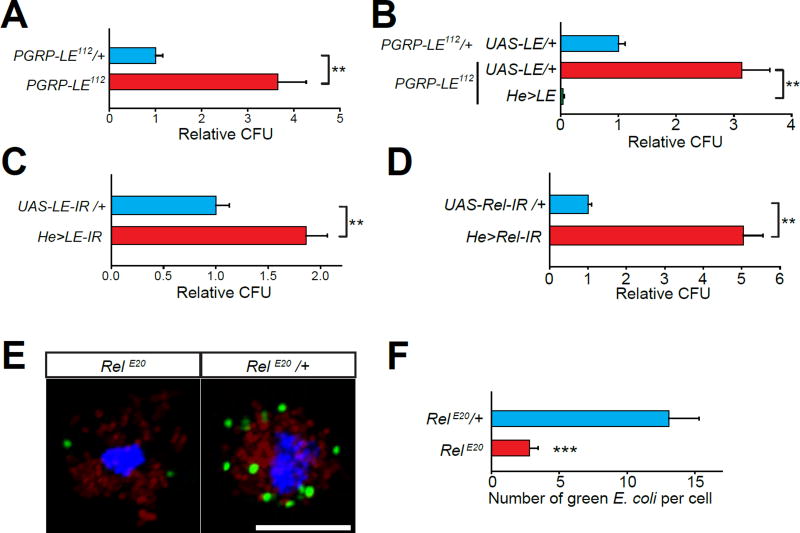
Figure 5. PGRP-LE and Relish signaling are necessary for macrophage mediated clearance of bacteria.
(A to D) CFU counts from flies of the indicated genotypes. (A) PGRP-LE null (PGRP-LE112) and control (PGRP-LE112/+) flies. (B) Macrophage overexpression of PGRP-LE (He>LE) in PGRP-LE null flies. (C and D) RNAi mediated knockdown of PGRP-LE (C) or Relish (D) in macrophages of wild-type flies. All CFU values (mean ±SEM) are normalized to means of the respective control genotype (blue bar). (E) Z-stack projections of confocal images of isolated larval macrophages with internalized/adhered fluorescent E. coli (red and green) from the “ex vivo sequential phagocytosis assay”. Shown are single larval macrophages isolated from RelE20 mutants and RelE20/+ control animals. Cells were stained with DAPI (blue). Scale bar represents 10 µm. (F) Quantification of data shown in (E). All values shown represent mean ±SEM.