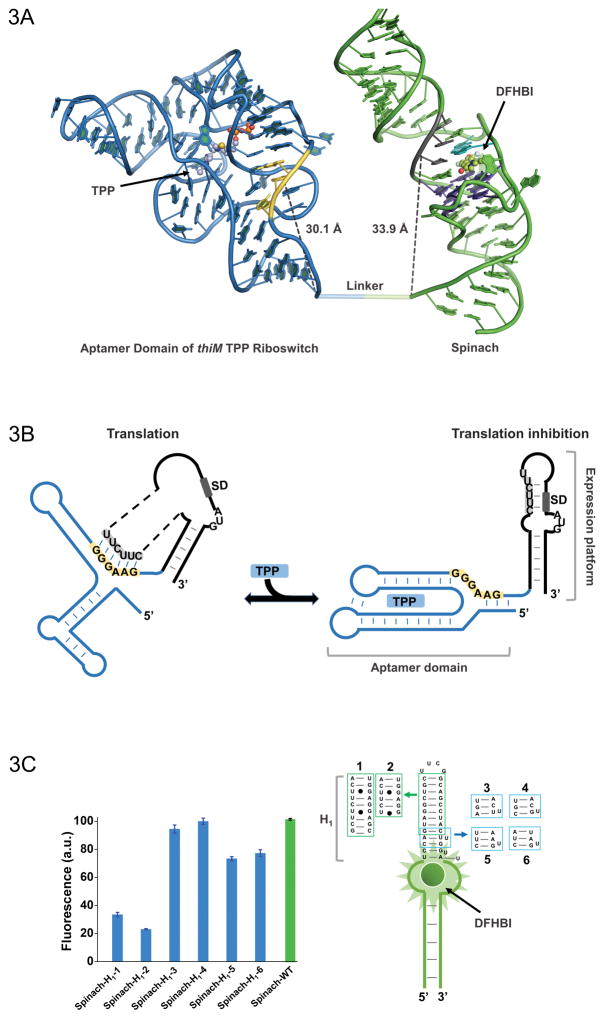
Figure 3.
(A) Model of the thiM-based Spinach riboswitch sensor for TPP. Riboswitch in blue with its switching sequence in yellow, while Spinach in green with transducer sequence in gray. Spinach U•A•U base triple is shown in cyan (overlap with most proximal residue of transducer sequence) and G-quadruplex shown in purple. TPP and DFHBI are shown in ball and stick representative. The distance between the switching sequence and transducer sequence to the base of thiM and Spinach, respectively, is quite similar, which would not be apparent in a two dimensional representation. (B) Mechanism of translational control by the thiM riboswitch in response to TPP. The transducer sequence shows complementarity to the 5′-AGGAGG-3′ Shine-Dalgarno sequence and switching sequence of the riboswitch’s aptamer domain. (C) H1 stem of Spinach tolerates certain modifications with only modest fluorescence changes. Such mutants expand the modularity of the approach and the range of transducer sequences that can be used for Spinach-based sensor design.