Figure 3. Using analog-sensitive PARP-1 mutants to unambiguously identify the ADP-ribosylation targets of DNA-dependent PARPs.
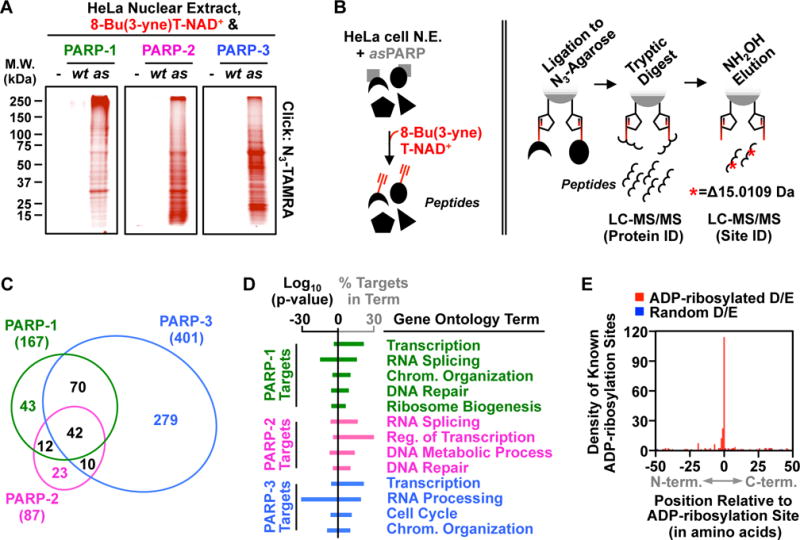
(A) In-gel fluorescence of HeLa cell nuclear extract proteins conjugated to azido-TAMRA following reactions with 8-Bu(3-yne)T-NAD+ in the presence of wild-type (wt) or analog-sensitive (as) PARP-1, PARP-2, or PARP-3.
(B) Depiction of the strategy for LC-MS/MS detection of PARP-specific ADP-ribosylation sites. (Left) asPARP-dependent labeling of HeLa cell nuclear extract (N.E.) proteins (represented by various shapes) using 8-Bu(3-yne)T-NAD+ (red). (Right) Post-labeling sample processing for LC-MS/MS. The 8-Bu(3-yne)T-ADP-ribosylated proteins are covalently linked to azide-agarose by copper-catalyzed cycloaddition (‘click’ chemistry; represented by pentagons), washed, and digested with trypsin to release peptides for protein identification. The remaining covalently linked peptides are eluted using hydroxyl amine (NH2OH) with a mass shift of 15.0109 Da, which allows for identification of ADP-ribosylation sites.
(C) Venn diagram depicting the overlap of the protein targets of PARP-1, PARP-2, and PARP-3.
(D) Gene ontology terms enriched for the sets of PARP-1, PARP-2 and PARP-3 targets.
(E) Histogram of the two-dimensional relationship between previously identified ADP-ribosylation sites (9) with those identified herein.
