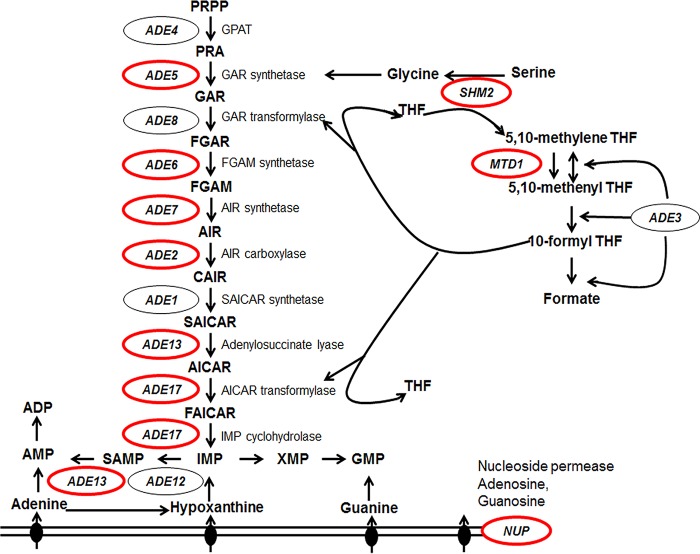
FIG 1 .
The purine salvage, AMP de novo biosynthesis, and one-carbon metabolic pathways of C. albicans. Genes identified by microarray analysis are circled in red. Enzymes and the genes encoding them that catalyze de novo purine biosynthesis and salvage pathways are as follows (from top to bottom): glutamine phosphoribosylpyrophosphate amidotransferase, ADE4; glycinamide ribotide synthase, ADE5; glycinamide ribotide transformylase, ADE8; formylglycinamide synthase, ADE6; aminoimidazole ribotide synthase, ADE7; aminoimidazole ribotide carboxylase, ADE2; succinylaminoimidazolecarboxamide ribotide synthase, ADE1; adenylosuccinate lyase, ADE13; aminoimidazole carboxamide ribotide transformylase and IMP cyclohydrolase, ADE17; adenylosuccinate synthase, ADE12; and nucleoside permease, NUP. Enzymes catalyzing the reactions in one-carbon metabolism and the genes that encode them are as follows: serine hydroxymethyltransferase, SHM2; NAD+-dependent 5,10-methylenetetrahydrafolate dehydrogenase, MTD1; mitochondrial C1-tetrahydrofolate synthase, ADE3 (MIS11). Intermediate metabolites are abbreviated as follows (from top to bottom): PRPP, 5-phosphoribosyl-α-1-pyrophosphate; PRA, 5-phospho-β-d-ribosylamine; GAR, 5-phosphoribosyl-glycinamide; FGAR, 5′-phosphoribosyl-N-formylglycinamide; FGAM, 5′-phosphoribosyl-N-formylglycinamidine; AIR, 5′-phosphoribosyl-5-aminoimidazole; CAIR, 5′-phosphoribosyl-5-amino-imidazole-4-carboxylate; SAICAR, 5-amino-4-imidazole-N-succinocarboxamide ribonucleotide; AICAR, 5-amino-4-imidazolecarboxamide ribonucleotide; FAICAR, 5′-phosphoribosyl-4-carboxamide-5-formamidoimidazole; IMP, inosine monophosphate; SAMP, adenylosuccinate; THF, tetrahydrofolate.