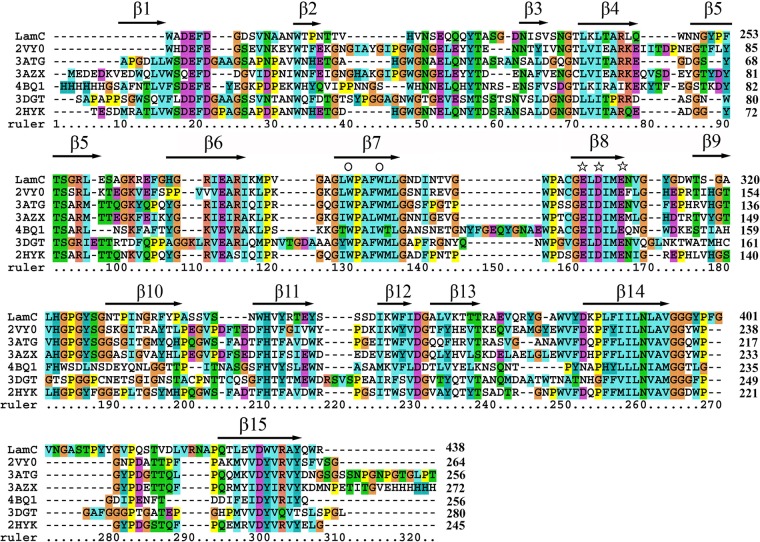
FIG 3.
Alignment of the GH16 domain sequence of the GH16 family β-glucanases. The proteins (and PDB numbers) were as follows: LamC from Corallococcus sp. EGB; PDB no. 3ATG, endo-β-(1,3)-glucanase from Cellulosimicrobium cellulans; PDB no. 2HYK, endo-β-(1,3)-glucanase of from Nocardiopsis sp. strain F96; PDB no. 4BQ1, ZgLamA from Z. galactanivorans; PDB no. 3AZX, laminarinase from T. maritima Msb8; PDB no. 2VY0, pfLamA from P. furiosus; and PDB no. 3DGT, endo-β-(1,3)-glucanase from S. sioyaensis. The protein sequence alignments were generated using the MUSCLE alignment in MEGA 7.0 (38). Secondary structures are labeled based on their appearance in 2HYK following a previous annotation (28). The stars identify the catalytic amino acids, including Glu-304, Asp-306, and Glu-309 (LamC numbering), and the conserved Trp residues are marked by the symbol ○ above the column. The numbers on the right are the amino acid residue positions in the whole sequence. The colors highlight other identities between sequences.