Figure 2.
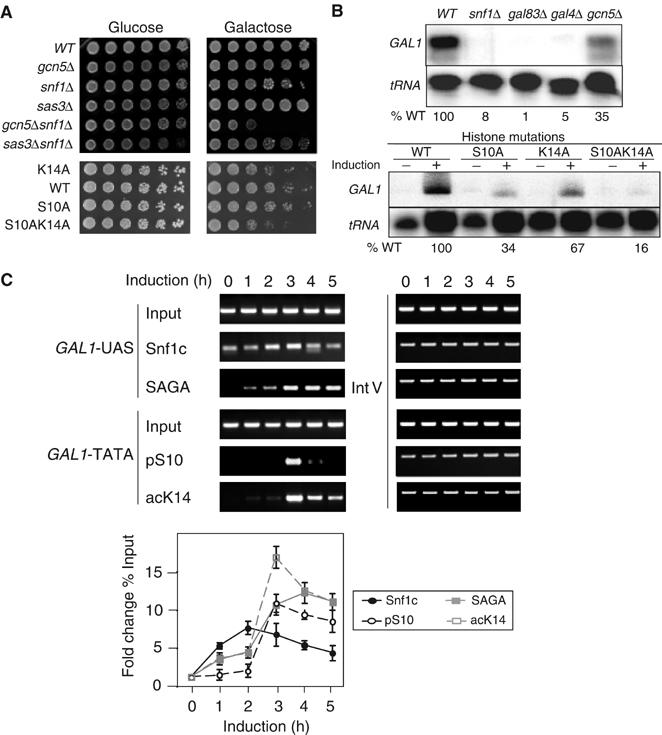
Cooperation of histone H3 phosphorylation and acetylation in galactose utilization and GAL1 transcriptional regulation in vivo. (A) Growth phenotype of enzyme deletions and histone substitutions. Five-fold serial dilutions of strains with the genotypes indicated on the left were spotted on glucose (2% glucose) and galactose (2% galactose) plates. Phenotypes were scored after 3-day growth at 30°C (top panels) and at 39°C (lower panels). (B) S1 nuclease RNA protection assay of enzyme deletions and histone substitutions. GAL1 transcription from WT, deletion strains (snf1, gal83, gal4, gcn5) and histone substitution strains (S10A, K14A, and S10AK14A) under repressing (−, 2% glucose) and inducing conditions (+, 2% galactose; upper panel). The induced level of GAL1 transcription in each strain was normalized to internal tRNA level and then calculated as percentage of WT level. (C) ChIP analysis of histone modifications on the GAL1 promoter during galactose induction time course. Cells were collected every 60 min over a 5 h time course. Antibodies were against Snf1 (Snf1c), pS10, Spt3-myc (SAGA), and acK14. Primer pairs were located in the GAL1 upstream activating sequence (GAL1-UAS) or the core promoter of GAL1 (GAL1-TATA).
