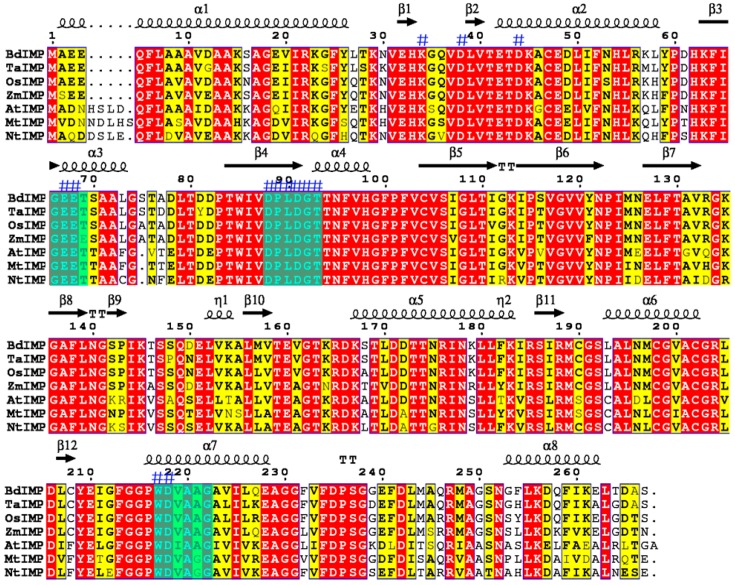
Figure 1.
Multiple sequence alignments of l-myo-inositol monophosphatase (IMPase) from other plant species. Flat figure shows the sequences of IMPase from several plants. The second structure element symbols presented on the top (squiggles are α helices, arrows β strands and TT letters beta turns), identical and similar residues are boxed in red and yellow, respectively. Three characteristic signature motifs (GEET, DPLDGT and WDXAAG) of the phosphatase super family are boxed by purple colours. The enzyme active sites (i.e., lysine[K]34, aspartate[D]38, 44, 88, 91, 218, glutamic acid[E]67, 68, proline[P]89, leucine[L]90, glycine[G]92, threonine [T]93 and tryptophan[W]217) are marked by blue “#”. BdIMP, Brachypodium distachyon IMP; TaIMP, Triticum aestivum IMP; OsIMP, Oryza sativa (Gaogonggui) IMP; ZmIMP, Zea mays; AtIMP, Arabidopsis thaliana IMP; MtIMP, Medicago truncatula IMP; NtIMP, Nicotiana tomentosiformis IMP.