Fig. 1.
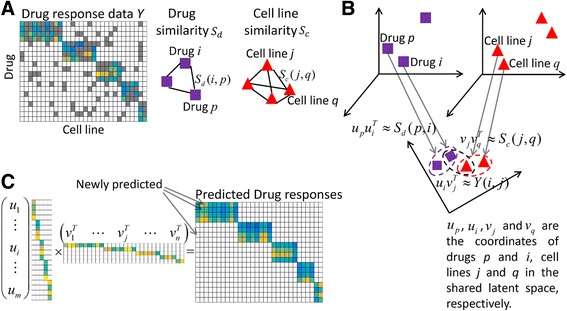
The framework of drug response prediction method SRMF. a The input data for SRMF includes the available drug responses (such as active area values) in cancer cell lines versus the unknown values marked as grey, chemical structure-based drug similarity and gene expression profile-based cell line similarity. b Rationale for the matrix factorization approach. Drugs and cell lines are mapped into a shared latent space with a low dimensionality. Furthermore, the associations among drugs and cell lines are described using the inner products of their coordinates in the shared latent space. c SRMF computes the coordinates of drugs and cell lines U and V in the shared latent space, which are used to reconstruct drug response matrix including the newly predicted drug responses
