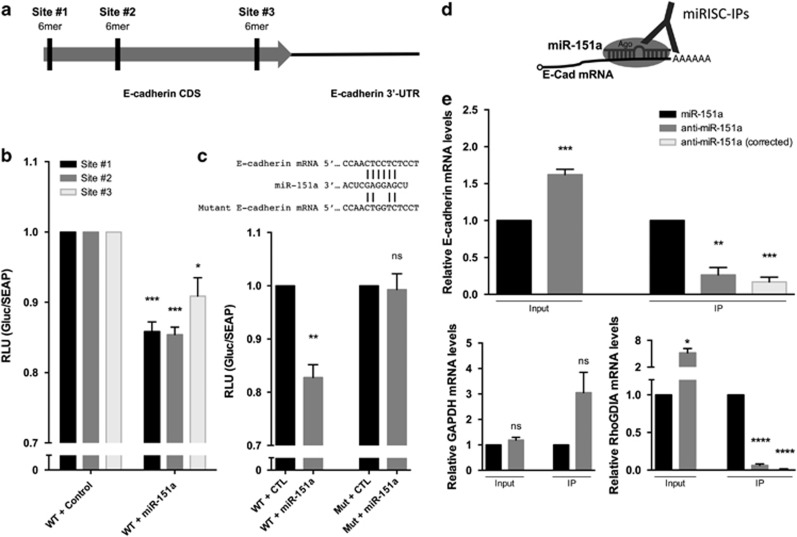
Figure 6.
miR-151a interacts with E-cadherin mRNA. (a) Schematic representation of three potential 6-mer miR-151a-binding sites in the coding DNA sequence (CDS) of E-cadherin mRNA. (b) Relative luciferase levels in A549 cells transfected with constructs expressing the wild type (WT) binding sequences at site #1, #2 or #3, along with miR-CTL or miR-151a mimics, were determined 48 hr after transfection. (n=3 independent cell cultures and experiments, 3 technical replicates of each). (c) Schematic of miR-151a binding to WT or mutant E-cadherin mRNA. Relative luciferase levels of A549 cells transfected with constructs expressing WT or mutated (site #2) binding sequences and miR-CTL or miR-151a mimics were determined 48 hr after transfection. (n=3 independent cell cultures and experiments, 3 technical replicates of each). (d) Schematic of Argonaute immunopurification strategy for miR-RNA complexes (Ago-RIP). (e) A549 cells stably expressing either miR-151a or anti-miR-151a were generated. Relative levels of E-cadherin, RhoGDIA and GAPDH mRNA in input and Ago-RIP (IP) fractions were determined. ‘Corrected’ indicates mRNA levels in IP fractions when normalized to the mRNA level in corresponding input fractions. (n=3 independent cell cultures and experiments, 3 technical replicates of each). Throughout figure, all graphs are shown as mean±s.e.m. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 by two-tailed Student’s t-test.