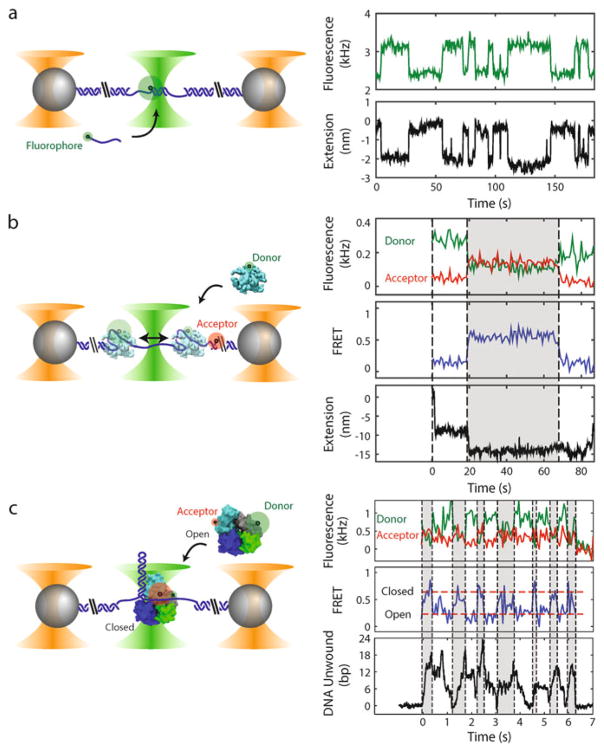
Fig. 1.
Examples of experiments with combined high-resolution optical trap and fluorescence (not to scale). Left panels: Polystyrene microspheres (grey) are held in optical traps (orange cones), tethered by an engineered DNA molecule (blue) containing a variable central segment flanked by long double-stranded DNA (dsDNA) handles. Fluorophores are excited by a green laser (green cone). Right panels: Time traces showing simultaneous measurement of fluorescence and tether extension. (a) Oligonucleotide hybridization. Short oligonucleotides (blue line) labeled with a fluorophore (green disk) bind and unbind to a complementary ssDNA section in the center of the tethered DNA. The fluorescence and change in tether extension upon hybridization are recorded simultaneously. (b) Single-stranded DNA binding protein wrapping dynamics. A tethered DNA molecule containing a short ssDNA region is labeled with a FRET acceptor at the ss-dsDNA junction (red disk). An E. coli single-stranded DNA binding protein (SSB, cyan) labeled with a FRET donor (green disk) binds to and wraps ssDNA around itself. Simultaneous measurement of FRET efficiency and tether extension enables determination of both the position of SSB along the tether and the amount of ssDNA wrapped. The SSB can transiently wrap and unwrap ssDNA under tension (e.g., t = 20 s), and can diffuse one-dimensionally along the ssDNA by reptation (t = 70 s) (data reproduced from ref. [22] with permission from eLife Sciences Publications). (c) UvrD helicase conformational and unwinding dynamics. A tethered DNA molecule contains a hairpin and short ssDNA protein loading site. E. coli UvrD helicase (cyan, blue, grey, and green) is labeled with FRET donor and acceptor pair to differentiate between two possible conformational states: “Open” and “Closed.” Simultaneous measurement of FRET efficiency and number of DNA base pairs of the hairpin unwound by the helicase enables correlation of the conformation with the activity of the helicase. Changes in UvrD conformational state correspond to switches between unwinding and rezipping of the DNA hairpin (reproduced from ref. [23] with permission from AAAS). The proteins in this figure were prepared with VMD [48] using PDB entries 1EYG, 2IS2, and 3LFU