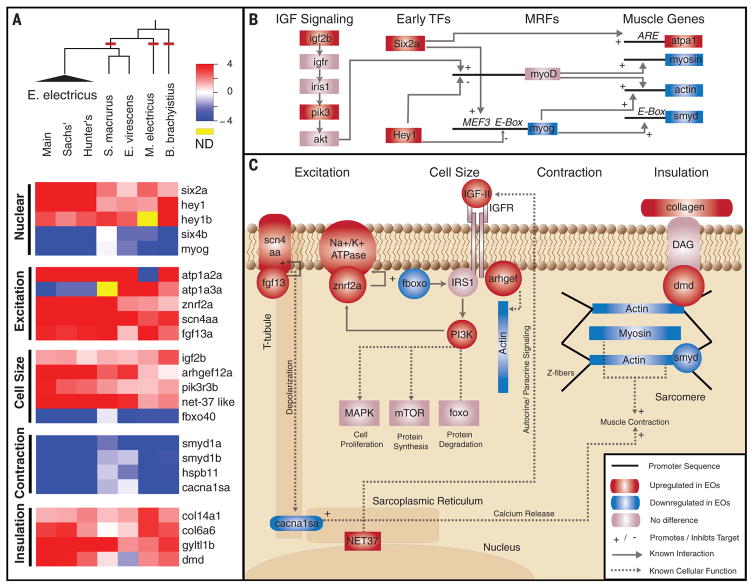
Fig. 2. Common toolkit for convergent evolution of EOs.
(A) RNA-Seq was performed on five species, representing three independent origins of electrogenesis (cladogram, red lines). Also shown are plots of the log-transformed ratio of EO to skeletal muscle expression genes (red, up-regulated in EO; blue, down-regulated in EO) in several categories of function, including (i) nuclear transcription factors, (ii) genes that regulate cell excitation, (iii) genes that regulate cell size, (iv) genes involved in contraction and excitation contraction coupling, and (v) genes encoding proteins that surround individual electrocytes to provide the scaffold for insulation. hey1b data for E. electricus was derived from the Trinity transcriptome assembly (6). (B) Interaction of identified IGF signaling and transcription factors (TFs). IGF signaling pathway genes and early TFs influence the expression of muscle regulatory factors (MRFs), which ultimately lead to the expression of muscle-specific effector genes (table S3). (C) Interactions of genes identified in (A) are shown, grouped by function. For each, we list known patterns of expression in electric fish or the result of knockout studies in other vertebrates (table S4). IGFR, IGF receptor; MAPK, mitogen-activated protein kinase.