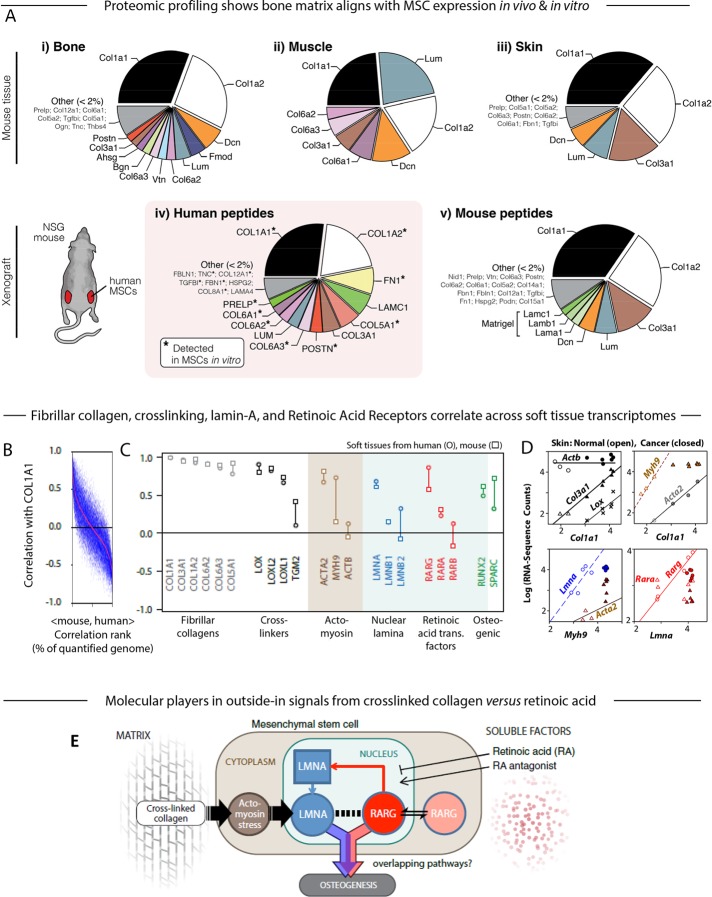
FIGURE 2:
Proteomic and transcriptomic profiles of mouse tissue and human xenografts suggest the key components in a model of the microenvironment-to-osteogenesis signaling cascade. (A) Whole mouse tissues (i, bone; ii, muscle; iii, skin) were profiled by MS proteomics and the ratios of mean ion currents used to estimate the fractional compositions of the respective extracellular matrices. In all cases, the matrix was dominated by collagen-I. Human MSCs were also engrafted into mouse flank, excised after 4 wk, and examined by MS. Subsequent analysis allowed separate identification and quantification of mouse and human peptides (Supplemental Figure S1A; Swift et al., 2013a,b). The human peptides in the xenograft (iv) showed some similarity to the endogenous bone profile, and most of the detectable matrix proteins were also present in preengraftment MSCs (asterisk). Mouse peptides in the xenograft (v) suggested comparison to skin, perhaps unsurprising, given the location of the graft, and also showed likely remnants of the Matrigel component of the initial injection. (B) Pearson correlation between primary matrix component COL1A1 mRNA and other genes quantified in soft tissues of mouse and human (genes with common annotation, n ≈ 15,000), sorted by the mean Pearson coefficient in mouse and human (red line). (C) Pearson correlation between COL1A1 and transcripts for fibrillar collagens, cross-linking enzymes, actomyosin cytoskeleton proteins, nuclear lamina proteins, RAR, and osteogenic transcription factors. Many of these key components were in the top few percent of correlations with collagen-I, as seen by comparison to Figure 2B. (D) RNA-sequencing data from mouse skin of normal or induced squamous cell carcinomas (SCCs; Friedrichs et al., 2007) revealing power-law relations between many of the factors analyzed in C. (E) Gene circuit model of how extracellular factors, mechanical (matrix composition and cross-linking) and molecular (soluble factors such as RA), can influence MSC osteogenesis. Matrix stiffness induces stress in the actomyosin network, which feeds into levels of nucleoskeletal protein LMNA (Buxboim et al., 2014); LMNA regulates its own transcript RARG, which in turn can be modulated by soluble agonists and antagonists to the RA pathway (Swift et al., 2013b). Both LMNA, through regulation of the SRF pathway (Ho et al., 2013; Swift et al., 2013b; Buxboim et al., 2014; Talwar et al., 2014), and the RA pathway conceivably contribute to an osteogenic endpoint, but the extent of overlap between these pathways is not known (Figure 1A). All experiments, n ≥ 3 (mean + SEM).