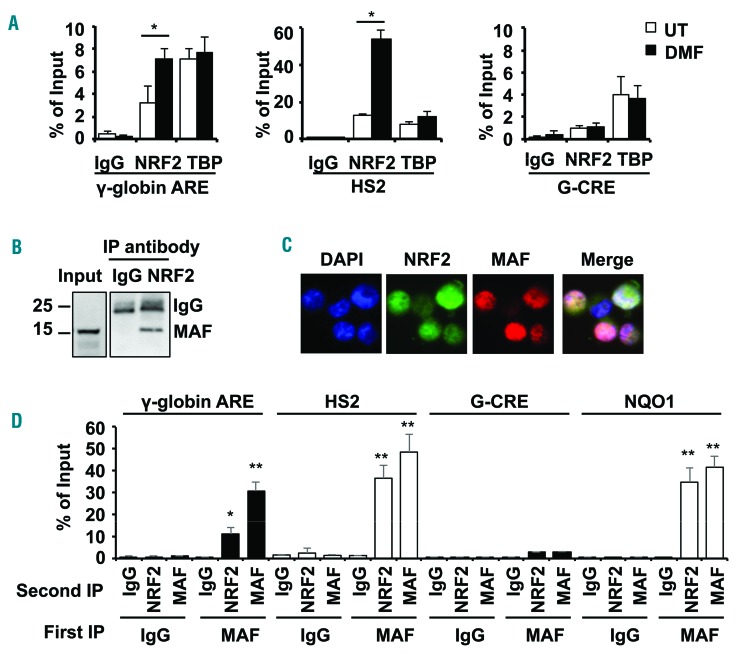
Figure 2.
NRF2/MAF heterodimers mediate γ-globin gene regulation. (A) ChIP assay was conducted with NRF2 and TATA binding protein (TBP) antibodies to determine in vivo binding in the γ-globin and HS2 ARE for untreated (white bars) and DMF treated (black bars) primary erythroid cells. The Gγ-globin cAMP response element (G-CRE) devoid of an ARE motif was used as a control region. The data are shown as the mean ± SEM of 3 independent experiments; *P<0.05. (B) Co-immunoprecipitation studies were completed to determine the interactions between NRF2 and MAF in whole cell protein lysates from KU812 leukemia cells treated with DMF (200 μM) for 48 hours. Immunoprecipitation (IP) with anti-NRF2 or IgG antibody was followed by western blot with anti-MAF antibody. (C) Cellular localization of NRF2 and MAF proteins was determined by immunofluorescence staining with rabbit anti-NRF2 and mouse anti-MAF antibodies followed by anti-rabbit secondary FITC antibody (green) and anti-mouse secondary phycoerythrine (PE) antibody (red) (Online Supplementary Methods). Localization of NRF2 and MAF in the nucleus was identified in the merged images; color code: MAF>NRF2, pink; NRF2>MAF, purple and NRF2=MAF, white. (D) Sequential-ChIP was conducted (Online Supplementary Methods) to determine if NRF2 and MAF co-localize at the regions shown in KU812 cells treated with DMF to stimulate NRF2 translocation to the nucleus. The G-CRE was used as a control region. Sequential immunoprecipitations with the first antibody (First IP) and second antibody (Second IP) were conducted; **P<0.01.