Figure 2.
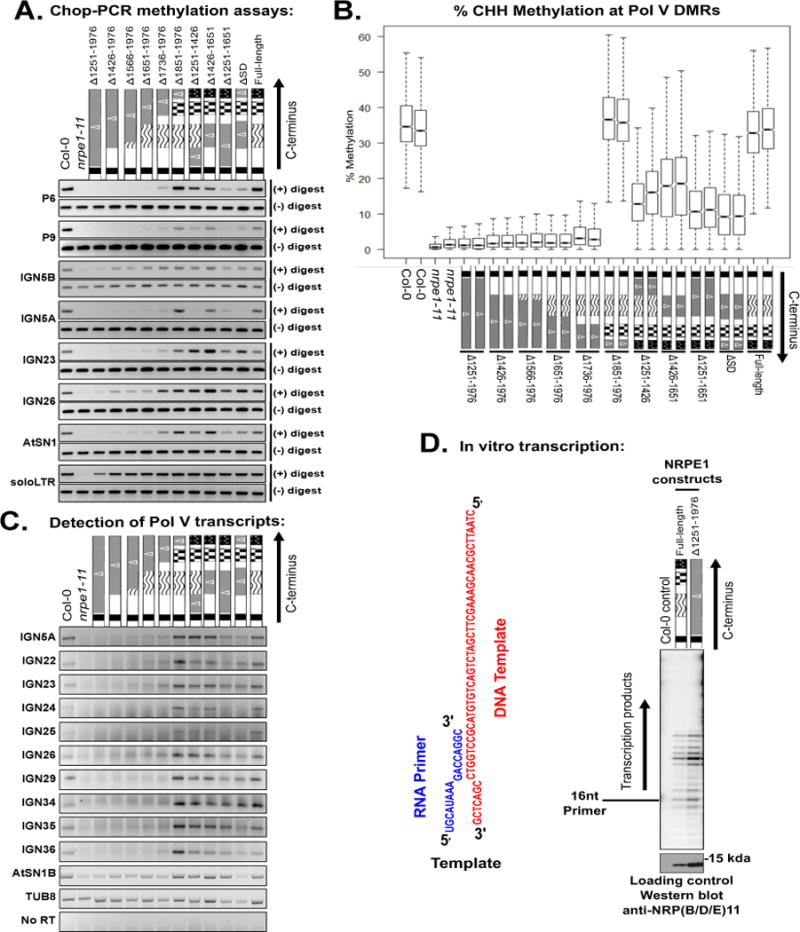
Effects of NRPE1 CTD deletions on CHH cytosine methylation and Pol V transcript abundance.
A. Chop-PCR assays conducted using AluI or HaeIII restriction endonucleases (see text for details). nrpe1-11 mutants expressing the indicated transgenes are compared to wild-type (Col-0) and nrpe1-11 controls. IGN5A and IGN5B adjacent regions, with A being intergenic and B corresponding to sequences internal to a LINE element targeted by RdDM (Wierzbicki et al., 2008).
B. Box plots displaying overall CHH cytosine methylation levels, determined by whole-genome bisulfite sequencing, within Pol V-dependent differentially methylated regions (DMRs).
C. RT-PCR detection of Pol V transcriptional products. TUB8 (Tubulin), transcribed by Pol II, serves as a control.
D. In vitro transcription activities of immunoprecipitated full length NRPE1 and NRPE1 missing the entire CTD on the indicated DNA template hybridized to an RNA primer. An immunoprecipitation fraction of non-transgenic Col-0 serves as a negative control. Relative protein input levels were compared by immunoblotting for the 11th subunit using anti-NRP(B/D/E)11. Transcription products are labeled by virtue of incorporation of 32P-CTP and visualized by phosphorimaging (Haag et al., 2012). See also Figures S2–S4, and Tables S1–S2.
