Figure 4.
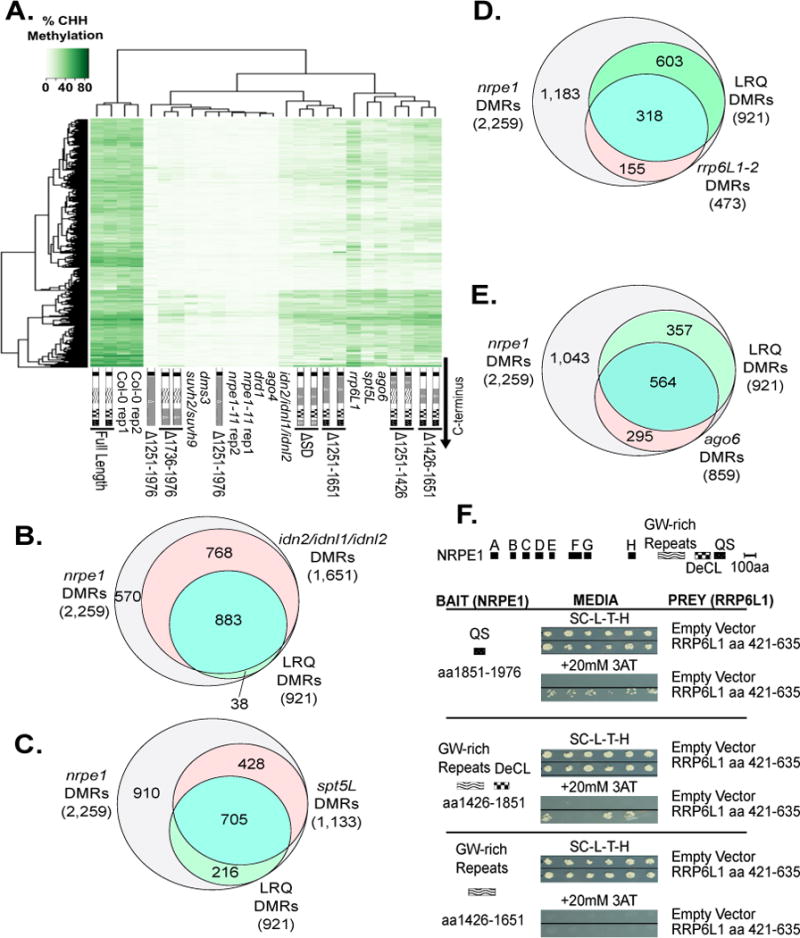
Relationship of methylation defects in Pol V CTD mutants to other RdDM mutants.
A. Heatmap clustering of % CHH methylation at Pol V DMRs, comparing CTD mutants, other RdDM pathway mutants (suvh2 suvh9, dms3, drd1, ago4, ago6, idn2 idnl1 idnl2, rrp6L1, spt5), and wild-type (Col-0) and nrpe1-11 controls.
B–E. Venn diagrams comparing total Pol V CHH DMRs, and LRQ DMRs in combination with DMRs dependent on idn2/idnl1/idnl2 (B), spt5L (C), rrp6L1 (D), or ago6 (E).
F. Yeast two hybrid (Y2H) interaction tests for Pol V CTD subdomains (“Bait”) and RRP6L1 aa 426–635 (“Prey”). Each bait was also tested against empty vector (pDEST22) controls. SC-L-T-H refers to media lacking leucine, tryptophan and histidine (top panel). Bait-prey interaction allows growth in the presence of 20 mM E-Amino-1, 2, 4 Triazol (3-AT), a HIS3 inhibitor (bottom panel). See also Figure S5 and Tables S1 and S2.
