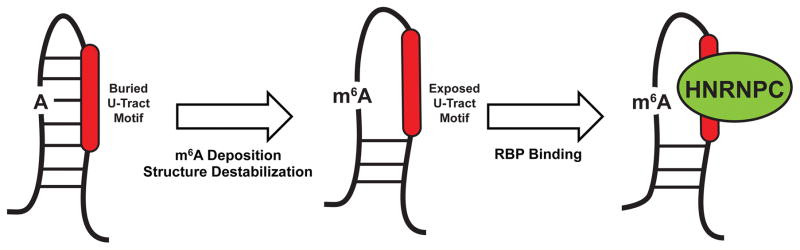
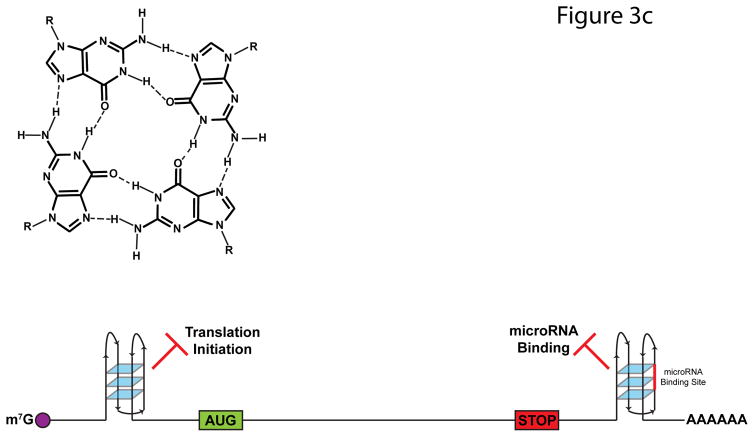
FIGURE 3. Impact of RNA Structures on Alternative Splicing.
a | A majority of RNA binding protein, Fox 2 homolog (RBFOX2) binding sites are over 500 nucleotides away from the nearest exon. To allow splicing regulation from these distal binding sites, base-pairing interactions occur within the intron, which bring RBFOX2 into close proximity to its target exons. b | N6-methyladenosine (m6A) deposition destabilizes base-pairing interactions and disrupts local secondary structure. In doing so, m6A can expose previously buried RNA-binding motifs to RNA-binding proteins, such as the U-tracts bound by heterogeneous ribonucleoprotein C (HNRNPC). The figure is reproduced, with permission, from REF. 80. c | G-qaudruplexes (RGQs) form from the stacking of three or more G-quartets (top). In 5′ untranslated regions (UTRs), RGQs impede the initiation and scanning steps of translation. In 3′UTRs, RGQs can block access to microRNA (miRNA) binding sites (shown in red), thus preventing miRNA-mediated decay and translational repression. These structures are dynamic and can be unwound by RNA helicases.