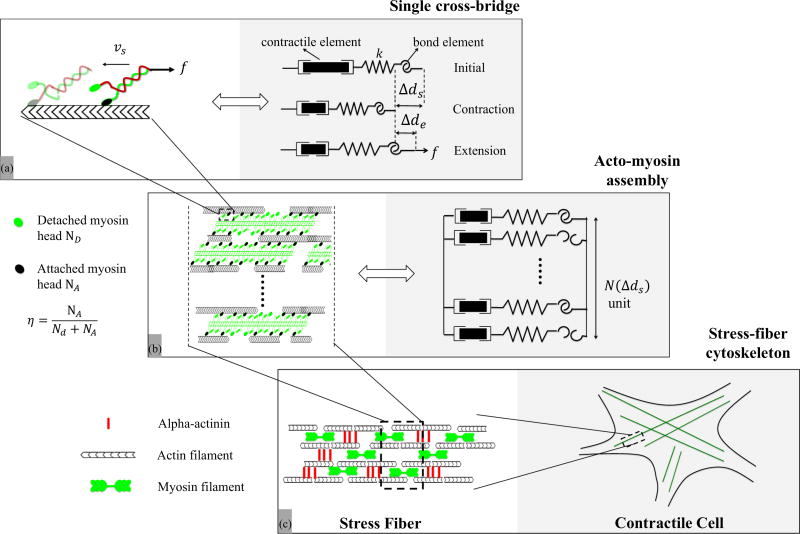
FIG. 1.
Overview of the contractile machinery of adherent cells from the molecular to the micron scale. (a) At the molecular scale, the dynamics of contraction starts from the motion of myosin motors that can attach to actin filaments and walk along them via coordinated configurational changes of the myosin head powered by ATP. The stability of these cross bridges is affected by the applied load f. In this study, this unit is modeled by a series of three elements comprised of a contractile element (whose contraction rate is υs), a compliant element of stiffness k, that captures the elasticity of the cross bridge and an “adhesive” element that represents the bond between actin and myosin. (b) A SF segment is primarily made of aligned and polarized actin filaments cross linked by a series of thick actin myofilaments whose motion along the actin tracks is facilitated by the motion of cross bridges. The overall SF contraction strain and force depend on the force generated by each individual myosin head and the number of attached actomyosin bonds, whose fraction is represented by the variable η. This organization can be represented by a parallel assembly of single cross-bridge elements shown in (a) which may be in an attached and detached state. (c) Assembled in series, these segments make up SFs which constitute the main contractile element of adherent cells. SFs typically organize into a well aligned network whose elements can span a cell between two adhesion points. υs is the sliding velocity, f is the external force against contraction, k is the actomyosin bond stiffness, Δds is the contraction, Δde is the elastic stretch of the bond, η is the ratio of the number of attached cross bridges to that of total available cross bridges N(Δds) at contraction Δds.