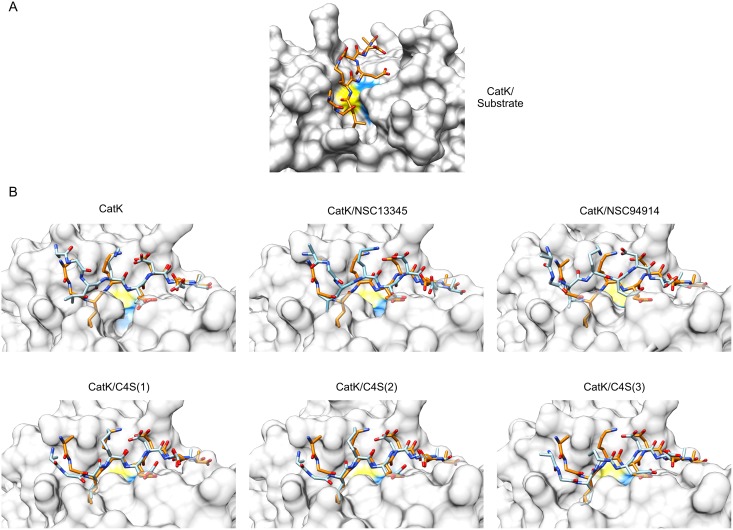
Fig 6. Docking of the substrate AGLKEDDA into the active sites of average MD conformers of cathepsin K.
(A) Best docking solution of the substrate AGLKEDDA (orange-themed sticks) bound to the average MD conformer from the simulation of the enzyme/substrate complex which illustrates productive binding of the substrate into the active site. (B) Best solutions of docking of AGLKEDDA to the average MD conformers from simulations of free cathepsin K, in complexes with small molecule effectors NSC13345 and NSC94914, and in complexes with C4S bound to each of the three known binding sites, respectively. The enzyme is shown in a transparent surface representation. The docked substrate is shown as blue-themed sticks. For comparison, the productively bound substrate molecule from panel A (orange-themed sticks) was superposed onto each complex. In both panels, the positions of active site Cys and His residues are colored yellow and blue, respectively. Molecular graphics were prepared with UCSF CHIMERA software [36].