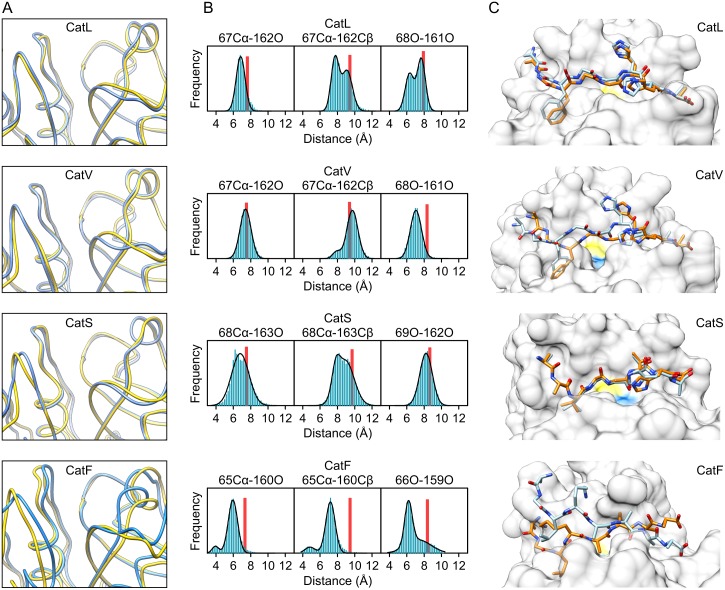
Fig 7. Conformational variability of the active sites of other human papain-like endopeptidases.
(A) Superpositions of initial X-ray structures (yellow ribbons) and average MD conformers (blue ribbons) of human cathepsins L, V, S and F (PDB accession codes 1MHW, 1FH0, 2OP3 and 1M6D, respectively). (B) Interatomic distances between residues lining the S2-S1 cleft in each peptidase. All populations were segregated into 0.2 Å wide bins. The curves were fitted assuming normal distribution of interatomic distances with models assuming one or two populations, as appropriate. Distances observed in initial X-ray structures are represented by red bars. The analyses were performed with Graph Pad Prism 5.0 Software. (C) Best solutions of docking substrate molecules into the active sites of cathepsins L, V, S and F. The proteins are shown in surface representation and the positions of catalytic Cys and His residues are colored yellow and blue, respectively. For each enzyme, solutions obtained with the initial X-ray structure and the average MD conformer are shown in superposition and the docked substrates are colored in orange and blue themes, respectively. The substrates used were AGFGGHHA for cathepsins L and V, AAVGGTHA for cathepsin S and AGLKAAAA for cathepsin F. Molecular graphics were prepared with UCSF CHIMERA software [36].