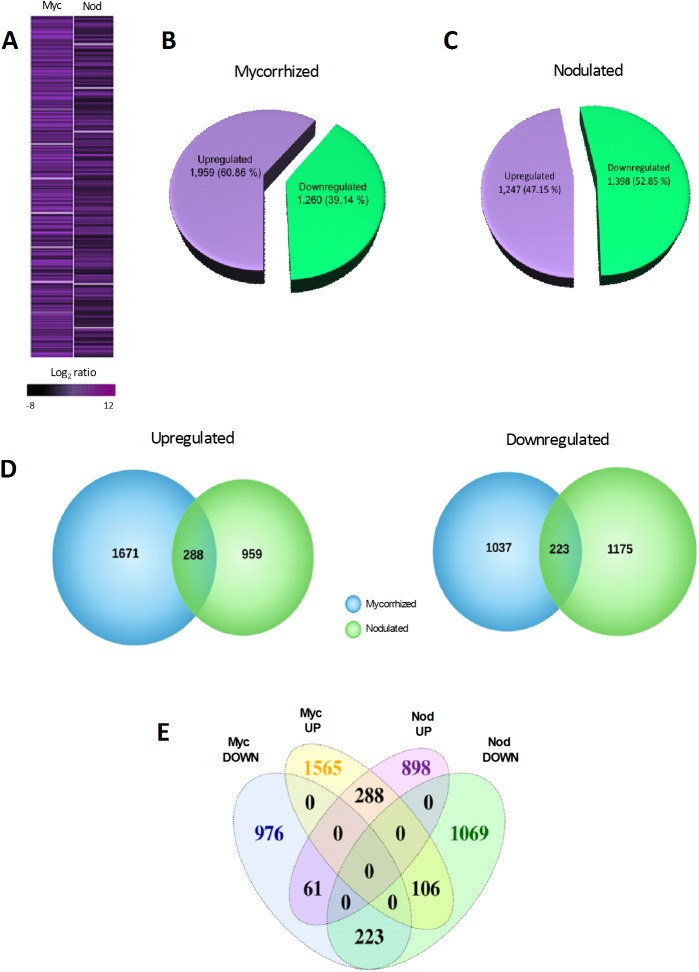
Fig 2. Summary of genome-wide expression profiling and identified DEGs in response to mycorrhizal and rhizobial colonization in P. vulgaris roots.
Global transcriptome profile of genes upregulated and downregulated in response to arbuscular mycorrhizal fungi and rhizobia (unpaired Student´s t-test, P-values of ≤0.05 and fold-change of ≥2.0 upregulated and downregulated) (A). Number and percent distribution of uniquely expressed upregulated and downregulated genes during (B) mycorrhizal and (C) nodulation conditions. The P. vulgaris locus names and descriptions are listed in S3 Table. Venn diagram showing (D) shared DEGs (left side, upregulated genes; right side, downregulated genes) and (E) number of overlapping DEGs in upregulated and downregulated mycorrhized and nodulated roots (clustered into four comparison groups represented by four ellipses).