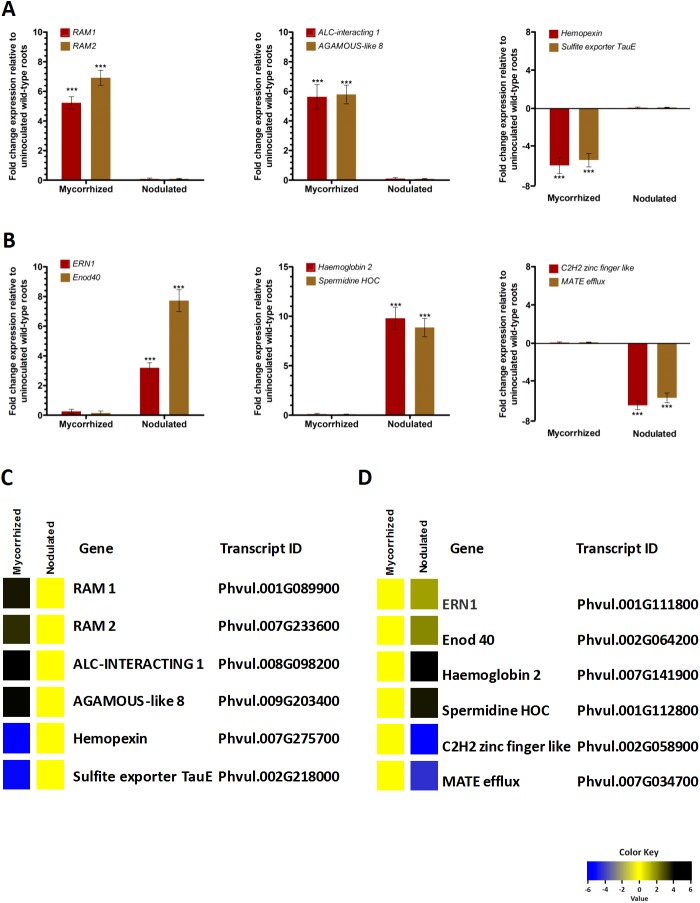
Fig 4. Validation of expression patterns of 12 DEGs of symbiont-colonized P. vulgaris roots by RT-qPCR analysis.
Twelve DEGs (differentially expressed genes) showed similar expression patterns between RT-qPCR data (A and B) and RNA-Seq data (C and D). These genes included four genes well characterized mycorrhizal symbiosis specific RAM1 and RAM2, nodule specific ERN1 and ENOD40; Four genes ALC-INTERACTING 1, AGAMOUS-LIKE 8 and HEMOPEXIN, SULFITE EXPORTER TAU-E that are up- and down-regulated under mycorrhization, respectively; two genes HAEMOGLOBIN 2, SPERMIDINE HOC and C2H2 ZINC FINGER LIKE, MATE EFFLUX up- and down-regulated under nodulation, respectively. RT-qPCR data are the averages of three biological replicates (n>9). Statistical significance of differences between mycorrhized and nodulated roots was determined using an unpaired two-tailed Student’s t-test (***P<0.001). Error bars represent means ± SEM.