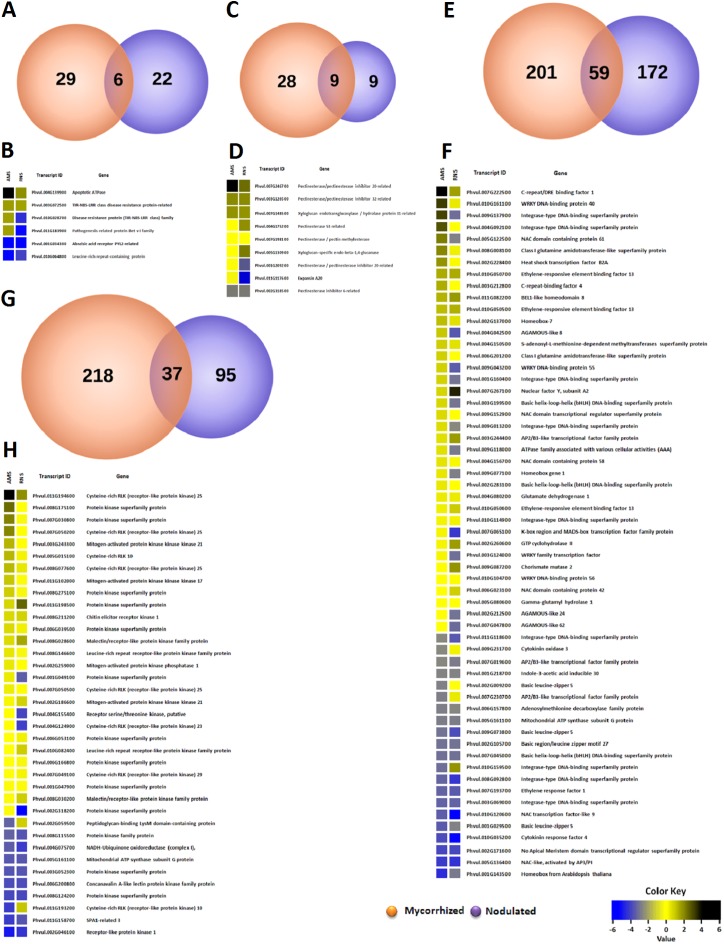
Fig 5. RNA-Seq based transcriptome comparison of genes induced by AMF and rhizobia in P. vulgaris roots.
Differentially expressed genes upregulated and downregulated in response to AMF and rhizobia in P. vulgaris roots were identified based on P-values of ≤0.05 and fold-change of ≥2.0 (upregulated and downregulated). Unique and shared genes were identified after a pairwise comparison between treatments. Venn diagrams representing the number of DEGs involved in processes related to (A) defense, (C) cell wall, (E) nitrogen metabolism, and (G) phosphate metabolism. Heat maps of the overlapping gene expression patterns during mycorrhizal and rhizobial colonization for processes related to (B) defense, (D) cell wall, (F) nitrogen metabolism, and (H) phosphate metabolism. Color bar shows the fold-change range, with red and green representing downregulation and upregulation, respectively.