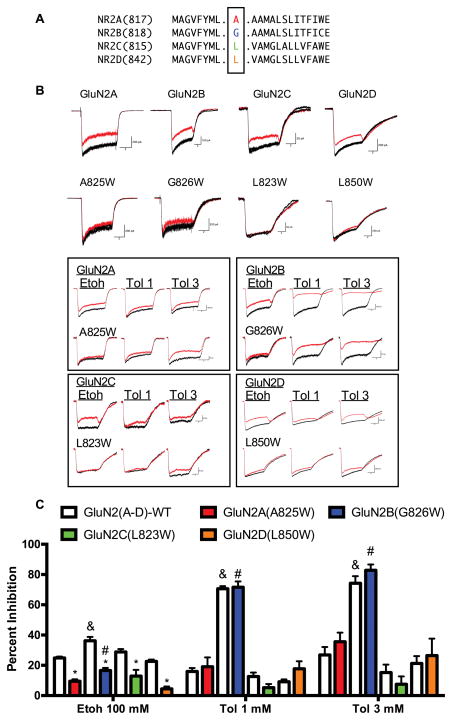
Figure 1.
Effect of TM4 tryptophan mutations on ethanol and toluene inhibition of recombinant NMDARs expressed in HEK293 cells. Cells were transfected with the GluN1-1a subunit and either wild-type or mutant GluN2(A–D) subunits. (A) Sequence alignment of TM4 domain of rat GluN2 subunits. Residues mutated to tryptophan (W) are highlighted in color. (B) Exemplar traces from wild-type and mutant NMDARs currents in the absence (black traces are control and washout) and presence (red trace) of 100 mM ethanol or toluene (1 mM, 3 mM). Scale bars: Upper traces: x-axis 2 sec; y-axis: GluN2A (200 pA), A825W (200 pA); GluN2B (100 pA), G826W (200 pA); GluN2C (50 pA), L823W (50 pA); GluN2D (200 pA), L850W (200 pA). Lower traces (boxed): x-axis 2 sec; y-axis: GluN2A (100 pA), A825W (50 pA); GluN2B (100 pA), G826W (50 pA); GluN2C (50 pA), L823W (50 pA); GluN2D (200 pA), L850W (200 pA). (C) Summary plot showing percent inhibition of steady-state currents by 100 mM ethanol or toluene (1 mM, 3 mM). Data are mean ± SEM (N: GluN2A (43), A825W (10); GluN2B (6), G826W (6); GluN2C (9), L823W (7); GluN2D (4), L850W (4). Symbols: &, value for GluN2B significantly different from GluN2A and GluN2D (Etoh p < 0.004); or GluN2A, 2C and 2D subunits (Toluene p < 0.0001); #value for GluN2B(G826W) significantly different from GluN2D(L850W) (Etoh p < 0.02) or GluN2A(A825W), 2C(L823W) and 2D(L850W) subunits (Toluene p < 0.0001). *: value for TM4-W mutant significantly different from corresponding wild-type control (p < 0.0001).